Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways
- PMID: 18256688
- PMCID: PMC2265754
- DOI: 10.1038/emboj.2008.17
Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways
Abstract
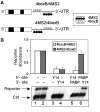
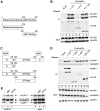
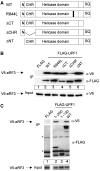
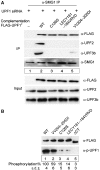
Nonsense-mediated mRNA decay (NMD) represents a key mechanism to control the expression of wild-type and aberrant mRNAs. Phosphorylation of the protein UPF1 in the context of translation termination contributes to committing mRNAs to NMD. We report that translation termination is inhibited by UPF1 and stimulated by cytoplasmic poly(A)-binding protein (PABPC1). UPF1 binds to eRF1 and to the GTPase domain of eRF3 both in its GTP- and GDP-bound states. Importantly, mutation studies show that UPF1 can interact with the exon junction complex (EJC) alternatively through either UPF2 or UPF3b to become phosphorylated and to activate NMD. On this basis, we discuss an integrated model where UPF1 halts translation termination and is phosphorylated by SMG1 if the termination-promoting interaction of PABPC1 with eRF3 cannot readily occur. The EJC, with UPF2 or UPF3b as a cofactor, interferes with physiological termination through UPF1. This model integrates previously competing models of NMD and suggests a mechanistic basis for alternative NMD pathways.
Figures


References
-
- Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV (2006) In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell 125: 1125–1136 - PubMed
-
- Altamura N, Groudinsky O, Dujardin G, Slonimski PP (1992) NAM7 nuclear gene encodes a novel member of a family of helicases with a Zn-ligand motif and is involved in mitochondrial functions in Saccharomyces cerevisiae. J Mol Biol 224: 575–587 - PubMed
-
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A (2004) A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432: 112–118 - PubMed
-
- Amrani N, Dong S, He F, Ganesan R, Ghosh S, Kervestin S, Li C, Mangus DA, Spatrick P, Jacobson A (2006) Aberrant termination triggers nonsense-mediated mRNA decay. Biochem Soc Trans 34: 39–42 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

