Molecular typing of Australian Scedosporium isolates showing genetic variability and numerous S. aurantiacum
- PMID: 18258122
- PMCID: PMC2600218
- DOI: 10.3201/eid1402.070920
Molecular typing of Australian Scedosporium isolates showing genetic variability and numerous S. aurantiacum
Erratum in
- Emerg Infect Dis. 2008 Apr;14(4):696
Abstract
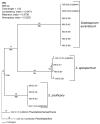
One hundred clinical isolates from a prospective nationwide study of scedosporiosis in Australia (2003-2005) and 46 additional isolates were genotyped by internal transcribed spacer-restriction fragment length polymorphism (ITS-RFLP) analysis, ITS sequencing, and M13 PCR fingerprinting. ITS-RFLP and PCR fingerprinting identified 3 distinct genetic groups. The first group corresponded to Scedosporium prolificans (n = 83), and the other 2 comprised isolates previously identified as S. apiospermum: one of these corresponded to S. apiospermum (n = 33) and the other to the newly described species S. aurantiacum (n = 30). Intraspecies variation was highest for S. apiospermum (58%), followed by S. prolificans (45%) and S. aurantiacum (28%) as determined by PCR fingerprinting. ITS sequence variation of 2.2% was observed among S. apiospermum isolates. No correlation was found between genotype of strains and their geographic origin, body site from which they were cultured, or colonization versus invasive disease. Twelve S. prolificans isolates from 2 suspected case clusters were examined by amplified fragment length polymorphism analysis. No specific clusters were confirmed.
Figures

References
-
- Steinbach WJ, Perfect JR. Scedosporium species infections and treatments. J Chemother. 2003;15(Suppl 2):16–27. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
