Structure-function analyses of single nucleotide polymorphisms in human N-acetyltransferase 1
- PMID: 18259988
- PMCID: PMC2265210
- DOI: 10.1080/03602530701852917
Structure-function analyses of single nucleotide polymorphisms in human N-acetyltransferase 1
Abstract
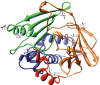
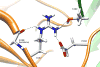
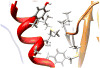
Human N-acetyltransferase 1 (NAT1) alleles are characterized by one or more single nucleotide polymorphisms (SNPs) associated with rapid and slow acetylation phenotypes. NAT1 both activates and deactivates arylamine drugs and carcinogens, and NAT1 polymorphisms are associated with increased frequencies of many cancers and birth defects. The recently resolved human NAT1 crystal structure was used to evaluate SNPs resulting in the protein substitutions R64W, V149I, R187Q, M205V, S214A, D251V, E261K, and I263V. The analysis enhances knowledge of NAT1 structure-function relationships, important for understanding associations of NAT1 SNPs with genetic predisposition to cancer, birth defects, and other diseases.
Figures

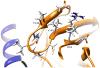



References
-
- Adam PJ, Berry J, Loader JA, Tyson KL, Craggs G, Smith P, De Belin J, Steers G, Pezzella F, Sachsenmeir KF, Stamps AC, Herath A, Sim E, O’Hare MJ, Harris AL, Terrett JA. Arylamine N-acetyltransferase-1 is highly expressed in breast cancers and conveys enhanced growth and resistance to etoposide in vitro. Mol Cancer Res. 2003;1:826–835. - PubMed
-
- Ambrosone CB, Abrams SM, Gorlewska-Roberts K, Kadlubar FF. Hair dye use, meat intake, and tobacco exposure and presence of carcinogen-DNA adducts in exfoliated breast ductal epithelial cells. Arch Biochem Biophys. 2007;464:169–175. - PubMed
-
- Bell DA, Stephens EA, Castranio T, Umbach DM, Watson M, Deakin M, Elder J, Hendrickse C, Duncan H, Strange RC. Polyadenylation polymorphism in the acetyltransferase 1 gene (NAT1) increases risk of colorectal cancer. Cancer Res. 1995;55:3537–3542. - PubMed
-
- Bieche I, Girault I, Urbain E, Tozlu S, Lidereau R. Relationship between intratumoral expression of genes coding for xenobiotic-metabolizing enzymes and benefit from adjuvant tamoxifen in estrogen receptor alpha-positive postmenopausal breast carcinoma. Breast Cancer Res. 2004;6:252–264. - PMC - PubMed
-
- Boukouvala S, Fakis G. Arylamine N-acetyltransferases: what we learn from genes and genomes. Drug Metab Rev. 2005;37:511–564. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
