Identification of molecular markers of bipolar cells in the murine retina
- PMID: 18260140
- PMCID: PMC2665264
- DOI: 10.1002/cne.21639
Identification of molecular markers of bipolar cells in the murine retina
Abstract
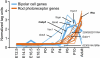
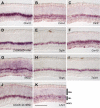
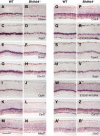
Retinal bipolar neurons serve as relay interneurons that connect rod and cone photoreceptor cells to amacrine and ganglion cells. They exhibit diverse morphologies essential for correct routing of photoreceptor cell signals to specific postsynaptic amacrine and ganglion cells. The development and physiology of these interneurons have not been completely defined molecularly. Despite previous identification of genes expressed in several bipolar cell subtypes, molecules that mark each bipolar cell type still await discovery. In this report, novel genetic markers of murine bipolar cells were found. Candidates were initially generated by using microarray analysis of single bipolar cells and mining of retinal serial analysis of gene expression (SAGE) data. These candidates were subsequently tested for expression in bipolar cells by RNA in situ hybridization. Ten new molecular markers were identified, five of which are highly enriched in their expression in bipolar cells within the adult retina. Double-labeling experiments using probes for previously characterized subsets of bipolar cells were performed to identify the subtypes of bipolar cells that express the novel markers. Additionally, the expression of bipolar cell genes was analyzed in Bhlhb4 knockout retinas, in which rod bipolar cells degenerate postnatally, to delineate further the identity of bipolar cells in which novel markers are found. From the analysis of Bhlhb4 mutant retinas, cone bipolar cell gene expression appears to be relatively unaffected by the degeneration of rod bipolar cells. Identification of molecular markers for the various subtypes of bipolar cells will lead to greater insights into the development and function of these diverse interneurons.
(c) 2008 Wiley-Liss, Inc.
Figures


References
-
- Baas D, Bumsted KM, Martinez JA, Vaccarino FM, Wikler KC, Barnstable CJ. The subcellular localization of Otx2 is cell-type specific and developmentally regulated in the mouse retina. Brain Res Mol Brain Res. 2000;78:26–37. - PubMed
-
- Baehr W, Champagne MS, Lee AK, Pittler SJ. Complete cDNA sequences of mouse rod photoreceptor cGMP phosphodiesterase alpha- and beta-subunits, and identification of beta′-, a putative beta-subunit isozyme produced by alternative splicing of the beta-subunit gene. FEBS Lett. 1991;278:107–114. - PMC - PubMed
-
- Bascom RA, Manara S, Collins L, Molday RS, Kalnins VI, McInnes RR. Cloning of the cDNA for a novel photoreceptor membrane protein (rom-1) identifies a disk rim protein family implicated in human retinopathies. Neuron. 1992;8:1171–1184. - PubMed
-
- Berrebi AS, Oberdick J, Sangameswaran L, Christakos S, Morgan JI, Mugnaini E. Cerebellar Purkinje cell markers are expressed in retinal bipolar neurons. J Comp Neurol. 1991;308:630–649. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

