Macrophage and T cell dynamics during the development and disintegration of mycobacterial granulomas
- PMID: 18261937
- PMCID: PMC2390753
- DOI: 10.1016/j.immuni.2007.12.010
Macrophage and T cell dynamics during the development and disintegration of mycobacterial granulomas
Abstract
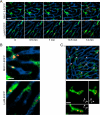
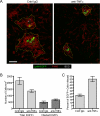
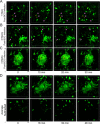
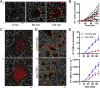
Granulomas play a key role in host protection against mycobacterial pathogens, with their breakdown contributing to exacerbated disease. To better understand the initiation and maintenance of these structures, we employed both high-resolution multiplex static imaging and intravital multiphoton microscopy of Mycobacterium bovis BCG-induced liver granulomas. We found that Kupffer cells directly capture blood-borne bacteria and subsequently nucleate formation of a nascent granuloma by recruiting both uninfected liver-resident macrophages and blood-derived monocytes. Within the mature granuloma, these myeloid cell populations formed a relatively immobile cellular matrix that interacted with a highly dynamic effector T cell population. The efficient recruitment of these T cells was highly dependent on TNF-alpha-derived signals, which also maintained the granuloma structure through preferential effects on uninfected macrophage populations. By characterizing the migration of both innate and adaptive immune cells throughout the process of granuloma development, these studies provide a new perspective on the cellular events involved in mycobacterial containment and escape.
Figures


Comment in
-
"The very pulse of the machine": the tuberculous granuloma in motion.Immunity. 2008 Feb;28(2):146-8. doi: 10.1016/j.immuni.2008.01.002. Immunity. 2008. PMID: 18275827
References
-
- Abadie V, Badell E, Douillard P, Ensergueix D, Leenen PJ, Tanguy M, Fiette L, Saeland S, Gicquel B, Winter N. Neutrophils rapidly migrate via lymphatics after Mycobacterium bovis BCG intradermal vaccination and shuttle live bacilli to the draining lymph nodes. Blood. 2005;106:1843–1850. - PubMed
-
- Algood HM, Chan J, Flynn JL. Chemokines and tuberculosis. Cytokine Growth Factor Rev. 2003;14:467–477. - PubMed
-
- Auffray C, Fogg D, Garfa M, Elain G, Join-Lambert O, Kayal S, Sarnacki S, Cumano A, Lauvau G, Geissmann F. Monitoring of blood vessels and tissues by a population of monocytes with patrolling behavior. Science. 2007;317:666–670. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

