A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis
- PMID: 18263776
- PMCID: PMC2276435
- DOI: 10.1105/tpc.107.056754
A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis
Abstract
Chitin, a polymer of N-acetyl-d-glucosamine, is found in fungal cell walls but not in plants. Plant cells can perceive chitin fragments (chitooligosaccharides) leading to gene induction and defense responses. We identified a LysM receptor-like protein (LysM RLK1) required for chitin signaling in Arabidopsis thaliana. The mutation in this gene blocked the induction of almost all chitooligosaccharide-responsive genes and led to more susceptibility to fungal pathogens but had no effect on infection by a bacterial pathogen. Additionally, exogenously applied chitooligosaccharides enhanced resistance against both fungal and bacterial pathogens in the wild-type plants but not in the mutant. Together, our data indicate that LysM RLK1 is essential for chitin signaling in plants (likely as part of the receptor complex) and is involved in chitin-mediated plant innate immunity. The LysM RLK1-mediated chitin signaling pathway is unique, but it may share a conserved downstream pathway with the FLS2/flagellin- and EFR/EF-Tu-mediated signaling pathways. Additionally, our work suggests a possible evolutionary relationship between the chitin and Nod factor perception mechanisms due to the similarities between their potential receptors and between the signal molecules perceived by them.
Figures
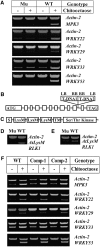
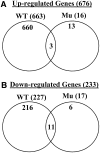


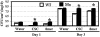
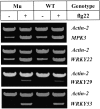
Comment in
-
Chitin signaling in plants: insights into the perception of fungal pathogens and rhizobacterial symbionts.Plant Cell. 2008 Feb;20(2):241-3. doi: 10.1105/tpc.108.058784. Plant Cell. 2008. PMID: 18285511 Free PMC article. No abstract available.
References
-
- Arlorio, M., Ludwig, A., Boller, T., and Bonafonte, P. (1992). Inhibition of fungal growth by plant chitinases and b-1,3-glucanases: A morphological study. Protoplasma 171 34–43.
-
- Asai, T., Tena, G., Plotnikova, J., Willmann, M.R., Chiu, W.L., Gomez-Gomez, L., Boller, T., Ausubel, F.M., and Sheen, J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415 977–983. - PubMed
-
- Baier, R., Schiene, K., Kohring, B., Flaschel, E., and Niehaus, K. (1999). Alfalfa and tobacco cells react differently to chitin oligosaccharides and Sinorhizobium meliloti nodulation factors. Planta 210 157–164. - PubMed
-
- Bateman, A., and Bycroft, M. (2000). The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD). J. Mol. Biol. 299 1113–1119. - PubMed
-
- Baureithel, K., Felix, G., and Boller, T. (1994). Specific, high affinity binding of chitin fragments to tomato cells and membranes: Competitive inhibition of binding by derivatives of chitooligosaccharides and a Nod factor of Rhizobium. J. Biol. Chem. 269 17931–17938. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

