Faecal ribosomal protein L19 is a genetic prognostic factor for survival in colorectal cancer
- PMID: 18266979
- PMCID: PMC4506161
- DOI: 10.1111/j.1582-4934.2008.00253.x
Faecal ribosomal protein L19 is a genetic prognostic factor for survival in colorectal cancer
Abstract
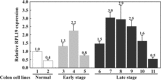
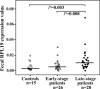
Ribosomal proteins are encoded by a gene family, members of which are overexpressed in human cancers. Many of them have been found, using oligonucleotide microarray hybridization, to be differentially expressed in the faeces of patients with various stages of colorectal cancer (CRC). The gene encoding ribosomal protein L19 (RPL19), a prognostic marker for human prostate cancer, is differentially expressed in CRC patients. Measurement of faecal RPL19 mRNA might improve prognostic prediction for CRC patients. Using quantitative real-time reverse transcription PCR, levels of RPL19 mRNA were detected in samples of colonic tissues from 44 CRC patients, in the faeces of 54 CRC patients and 15 controls, and in 11 colonic cell lines. Seven of 24 patients with late-stage CRC (Dukes' stages C and D) expressed over 2-fold more RPL19 in colonic tumour tissues than in corresponding normal tissues (P= 0.038). The mean faecal RPL19 mRNA levels of late-staged patients were higher than those of controls (P= 0.003) and early-staged patients (P= 0.008). Patients with both high serum levels of carcinoembryonic antigen (CEA; > 5 ng/mL) and high-faecal RPL19 mRNA (> or =0.0069) had higher risk (odds ratio, 8.0; P= 0.015) and lower overall 48-month survival (33.8 +/- 13.7%, P= 0.013). Oligonucleotide microarray hybridization analysis of faecal molecules identified gene transcripts differentially present in faeces. In conclusion, faecal RPL19 expression is associated with advanced tumour stages and addictive to serum CEA in predicting prognosis of CRC patients.
Figures


Similar articles
-
Ribosomal protein l19 is a prognostic marker for human prostate cancer.Clin Cancer Res. 2006 Apr 1;12(7 Pt 1):2061-5. doi: 10.1158/1078-0432.CCR-05-2445. Clin Cancer Res. 2006. PMID: 16609016
-
Factors that contribute to faecal cyclooxygenase-2 mRNA expression in subjects with colorectal cancer.Br J Cancer. 2010 Mar 2;102(5):916-21. doi: 10.1038/sj.bjc.6605564. Epub 2010 Feb 9. Br J Cancer. 2010. PMID: 20145612 Free PMC article.
-
Clinical significance of circulating tumor cells, including cancer stem-like cells, in peripheral blood for recurrence and prognosis in patients with Dukes' stage B and C colorectal cancer.J Clin Oncol. 2011 Apr 20;29(12):1547-55. doi: 10.1200/JCO.2010.30.5151. Epub 2011 Mar 21. J Clin Oncol. 2011. PMID: 21422427
-
The role of faecal calprotectin in diagnosis and staging of colorectal neoplasia: a systematic review and meta-analysis.BMC Gastroenterol. 2022 Apr 9;22(1):176. doi: 10.1186/s12876-022-02220-1. BMC Gastroenterol. 2022. PMID: 35397505 Free PMC article.
-
Biological and clinical markers in colorectal cancer: state of the art.Front Biosci (Schol Ed). 2010 Jan 1;2(2):422-31. doi: 10.2741/s75. Front Biosci (Schol Ed). 2010. PMID: 20036958 Review.
Cited by
-
Reciprocal regulation of Daxx and PIK3CA promotes colorectal cancer cell growth.Cell Mol Life Sci. 2022 Jun 19;79(7):367. doi: 10.1007/s00018-022-04399-8. Cell Mol Life Sci. 2022. PMID: 35718818 Free PMC article.
-
Downregulation of Methionine Cycle Genes MAT1A and GNMT Enriches Protein-Associated Translation Process and Worsens Hepatocellular Carcinoma Prognosis.Int J Mol Sci. 2022 Jan 1;23(1):481. doi: 10.3390/ijms23010481. Int J Mol Sci. 2022. PMID: 35008908 Free PMC article.
-
Ribosomal proteins: insight into molecular roles and functions in hepatocellular carcinoma.Oncogene. 2018 Jan 18;37(3):277-285. doi: 10.1038/onc.2017.343. Epub 2017 Sep 25. Oncogene. 2018. PMID: 28945227 Review.
-
Lowly expressed ribosomal protein s19 in the feces of patients with colorectal cancer.ISRN Gastroenterol. 2012;2012:394545. doi: 10.5402/2012/394545. Epub 2012 Jan 9. ISRN Gastroenterol. 2012. PMID: 22272377 Free PMC article.
-
The Oncogenic Effects, Pathways, and Target Molecules of JC Polyoma Virus T Antigen in Cancer Cells.Front Oncol. 2022 Mar 8;12:744886. doi: 10.3389/fonc.2022.744886. eCollection 2022. Front Oncol. 2022. PMID: 35350574 Free PMC article.
References
-
- Baak JP, Path FR, Hermsen MA, Meijer G, Schmidt J, Janssen EA. Genomics and proteomics in cancer. Eur J Cancer. 2003;39:1199–215. - PubMed
-
- Chin KV, Alabanza L, Fujii K, Kudoh K, Kita T, Kikuchi Y, Selvanayagam ZE, Wong YF, Lin Y, Shih WC. Application of expression genomics for predicting treatment response in cancer. Ann N Y Acad Sci. 2005;1058:186–95. - PubMed
-
- Cowell JK, Hawthorn L. The application of microarray technology to the analysis of the cancer genome. Curr Mol Med. 2007;7:103–20. - PubMed
-
- Fenech M. Biomarkers of genetic damage for cancer epidemiology. Toxicology. 2002;181-182:411–6. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

