Slipins: ancient origin, duplication and diversification of the stomatin protein family
- PMID: 18267007
- PMCID: PMC2258279
- DOI: 10.1186/1471-2148-8-44
Slipins: ancient origin, duplication and diversification of the stomatin protein family
Abstract
Background: Stomatin is a membrane protein that was first isolated from human red blood cells. Since then, a number of stomatin-like proteins have been identified in all three domains of life. The conservation among these proteins is remarkable, with bacterial and human homologs sharing 50 % identity. Despite being associated with a variety of diseases such as cancer, kidney failure and anaemia, precise functions of these proteins remain unclear.
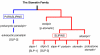
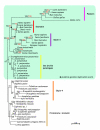
Results: We have constructed a comprehensive phylogeny of all 'stomatin-like' sequences that share a 150 amino acid domain. We show these proteins comprise an ancient family that arose early in prokaryotic evolution, and we propose a new nomenclature that reflects their phylogeny, based on the name "slipin" (stomatin-like protein). Within prokaryotes there are two distinct subfamilies that account for the two different origins of the eight eukaryotic stomatin subfamilies, one of which gave rise to eukaryotic SLP-2, renamed here "paraslipin". This was apparently acquired through the mitochondrial endosymbiosis and is widely distributed amongst the major kingdoms. The other prokaryotic subfamily gave rise to the ancestor of the remaining seven eukaryotic subfamilies. The highly diverged "alloslipin" subfamily is represented only by fungal, viral and ciliate sequences. The remaining six subfamilies, collectively termed "slipins", are confined to metazoa. Protostome stomatin, as well as a newly reported arthropod subfamily slipin-4, are restricted to invertebrate groups, whilst slipin-1 (previously SLP-1) is present in nematodes and higher metazoa. In vertebrates, the stomatin family expanded considerably, with at least two duplication events giving rise to podocin and slipin-3 subfamilies (previously SLP-3), with the retained ancestral sequence giving rise to vertebrate stomatin.
Conclusion: Stomatin-like proteins have their origin in an ancient duplication event that occurred early on in the evolution of prokaryotes. By constructing a phylogeny of this family, we have identified and named a number of orthologous groups: these can now be used to infer function of stomatin subfamilies in a meaningful way.
Figures

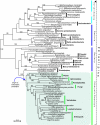
 indicates the clade uniting Chlorobi, Gammaproteobacteria and Spirochetes with two parallel phylogenies. Note the position of Rickettsiales as the sister group to the eukaryotic clade. Accession numbers available in Additional file 2.
indicates the clade uniting Chlorobi, Gammaproteobacteria and Spirochetes with two parallel phylogenies. Note the position of Rickettsiales as the sister group to the eukaryotic clade. Accession numbers available in Additional file 2.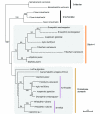


References
-
- Stewart G, Hepworth-Jones B, Keen J, Dash B, Argent A, Casimir C. Isolation of cDNA coding for an ubiquitous membrane protein deficient in high Na+, low K+ stomatocytic erythrocytes. Blood. 1992;79:1593–1601. - PubMed
-
- Wang D, Mentzer W, Cameron T, Johnson R. Purification of band 7.2b, a 31-kDa integral phosphoprotein absent in hereditary stomatocytosis. J Biol Chem. 1991;266:17826–17831. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

