A scenario on the stepwise evolution of the genetic code
- PMID: 18267295
- PMCID: PMC5054201
- DOI: 10.1016/S1672-0229(08)60001-7
A scenario on the stepwise evolution of the genetic code
Abstract
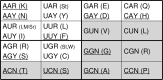
It is believed that in the RNA world the operational (ribozymes) and the informational (riboscripts) RNA molecules were created with only three (adenosine, uridine, and guanosine) and two (adenosine and uridine) nucleosides, respectively, so that the genetic code started uncomplicated. Ribozymes subsequently evolved to be able to cut and paste themselves and riboscripts were acceptive to rigorous editing (adenosine to inosine); the intensive diversification of RNA molecules shaped novel cellular machineries that are capable of polymerizing amino acids-a new type of cellular building materials for life. Initially, the genetic code, encoding seven amino acids, was created only to distinguish purine and pyrimidine; it was later expanded in a stepwise way to encode 12, 15, and 20 amino acids through the relief of guanine from its roles as operational signals and through the recruitment of cytosine. Therefore, the maturation of the genetic code also coincided with (1) the departure of aminoacyl-tRNA synthetases (AARSs) from the primordial translation machinery, (2) the replacement of informational RNA by DNA, and (3) the co-evolution of AARSs and their cognate tRNAs. This model predicts gradual replacements of RNA-made molecular mechanisms, cellular processes by proteins, and informational exploitation by DNA.
Figures



Similar articles
-
Emergence and evolution.Top Curr Chem. 2014;344:43-87. doi: 10.1007/128_2013_423. Top Curr Chem. 2014. PMID: 23478877 Free PMC article. Review.
-
Amino acid biogenesis, evolution of the genetic code and aminoacyl-tRNA synthetases.J Theor Biol. 2004 Jun 7;228(3):389-96. doi: 10.1016/j.jtbi.2004.01.014. J Theor Biol. 2004. PMID: 15135037
-
Four primordial modes of tRNA-synthetase recognition, determined by the (G,C) operational code.Proc Natl Acad Sci U S A. 1997 May 13;94(10):5183-8. doi: 10.1073/pnas.94.10.5183. Proc Natl Acad Sci U S A. 1997. PMID: 9144212 Free PMC article.
-
Origin and Evolution of the Universal Genetic Code.Annu Rev Genet. 2017 Nov 27;51:45-62. doi: 10.1146/annurev-genet-120116-024713. Epub 2017 Aug 30. Annu Rev Genet. 2017. PMID: 28853922 Review.
-
Symmetrical distributions of aminoacyl-tRNA synthetases during the evolution of the genetic code.Theory Biosci. 2023 Sep;142(3):211-219. doi: 10.1007/s12064-023-00394-0. Epub 2023 Jul 5. Theory Biosci. 2023. PMID: 37402895 Free PMC article.
Cited by
-
The pendulum model for genome compositional dynamics: from the four nucleotides to the twenty amino acids.Genomics Proteomics Bioinformatics. 2012 Aug;10(4):175-80. doi: 10.1016/j.gpb.2012.08.002. Epub 2012 Aug 11. Genomics Proteomics Bioinformatics. 2012. PMID: 23084772 Free PMC article.
-
Analysis of codon usage bias of envelope glycoprotein genes in nuclear polyhedrosis virus (NPV) and its relation to evolution.BMC Genomics. 2016 Aug 24;17(1):677. doi: 10.1186/s12864-016-3021-7. BMC Genomics. 2016. PMID: 27558469 Free PMC article.
-
Emergence and evolution.Top Curr Chem. 2014;344:43-87. doi: 10.1007/128_2013_423. Top Curr Chem. 2014. PMID: 23478877 Free PMC article. Review.
-
Modeling compositional dynamics based on GC and purine contents of protein-coding sequences.Biol Direct. 2010 Nov 8;5:63. doi: 10.1186/1745-6150-5-63. Biol Direct. 2010. PMID: 21059261 Free PMC article.
-
Randomness in Sequence Evolution Increases over Time.PLoS One. 2016 May 25;11(5):e0155935. doi: 10.1371/journal.pone.0155935. eCollection 2016. PLoS One. 2016. PMID: 27224236 Free PMC article.
References
-
- Singh S. Anchor Books; New York, USA: 1999. The Code Book.
-
- Gesteland R.F. second edition. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, USA: 1999. The RNA World: The Nature of Modern RNA Suggests a Prebiotic RNA.
-
- Joyce G.F. The rise and fall of the RNA world. New Biol. 1991;3:399–407. - PubMed
-
- Joyce G.F. The antiquity of RNA-based evolution. Nature. 2002;418:214–221. - PubMed
-
- Orgel L.E. The origin of life—a review of facts and speculations. Trends Biochem. Sci. 1998;23:491–495. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

