Apn1 and Apn2 endonucleases prevent accumulation of repair-associated DNA breaks in budding yeast as revealed by direct chromosomal analysis
- PMID: 18267974
- PMCID: PMC2346603
- DOI: 10.1093/nar/gkm1148
Apn1 and Apn2 endonucleases prevent accumulation of repair-associated DNA breaks in budding yeast as revealed by direct chromosomal analysis
Abstract
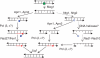
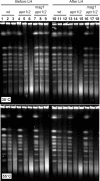
Base excision repair (BER) provides relief from many DNA lesions. While BER enzymes have been characterized biochemically, BER functions within cells are much less understood, in part because replication bypass and double-strand break (DSB) repair can also impact resistance to base damage. To investigate BER in vivo, we examined the repair of methyl methanesulfonate (MMS) induced DNA damage in haploid G1 yeast cells, so that replication bypass and recombinational DSB repair cannot occur. Based on the heat-lability of MMS-induced base damage, an assay was developed that monitors secondary breaks in full-length yeast chromosomes where closely spaced breaks yield DSBs that are observed by pulsed-field gel electrophoresis. The assay detects damaged bases and abasic (AP) sites as heat-dependent breaks as well as intermediate heat-independent breaks that arise during BER. Using a circular chromosome, lesion frequency and repair kinetics could be easily determined. Monitoring BER in single and multiple glycosylase and AP-endonuclease mutants confirmed that Mag1 is the major enzyme that removes MMS-damaged bases. This approach provided direct physical evidence that Apn1 and Apn2 not only repair cellular base damage but also prevent break accumulation that can result from AP sites being channeled into other BER pathway(s).
Figures




Similar articles
-
Deletion of the MAG1 DNA glycosylase gene suppresses alkylation-induced killing and mutagenesis in yeast cells lacking AP endonucleases.Mutat Res. 2001 Dec 19;487(3-4):137-47. doi: 10.1016/s0921-8777(01)00113-6. Mutat Res. 2001. PMID: 11738940
-
Involvement of two endonuclease III homologs in the base excision repair pathway for the processing of DNA alkylation damage in Saccharomyces cerevisiae.DNA Repair (Amst). 2004 Jan 5;3(1):51-9. doi: 10.1016/j.dnarep.2003.09.005. DNA Repair (Amst). 2004. PMID: 14697759
-
Alkylation base damage is converted into repairable double-strand breaks and complex intermediates in G2 cells lacking AP endonuclease.PLoS Genet. 2011 Apr;7(4):e1002059. doi: 10.1371/journal.pgen.1002059. Epub 2011 Apr 28. PLoS Genet. 2011. PMID: 21552545 Free PMC article.
-
Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae.DNA Repair (Amst). 2004 Jan 5;3(1):1-12. doi: 10.1016/j.dnarep.2003.10.002. DNA Repair (Amst). 2004. PMID: 14697754 Review.
-
Yeast base excision repair: interconnections and networks.Prog Nucleic Acid Res Mol Biol. 2001;68:29-39. doi: 10.1016/s0079-6603(01)68087-5. Prog Nucleic Acid Res Mol Biol. 2001. PMID: 11554305 Review.
Cited by
-
Increased chromosome mobility facilitates homology search during recombination.Nat Cell Biol. 2012 Apr 8;14(5):510-7. doi: 10.1038/ncb2472. Nat Cell Biol. 2012. PMID: 22484485
-
Deficient SUMO attachment to Flp recombinase leads to homologous recombination-dependent hyperamplification of the yeast 2 microm circle plasmid.Mol Biol Cell. 2009 Feb;20(4):1241-51. doi: 10.1091/mbc.e08-06-0659. Epub 2008 Dec 24. Mol Biol Cell. 2009. PMID: 19109426 Free PMC article.
-
Coincident resection at both ends of random, γ-induced double-strand breaks requires MRX (MRN), Sae2 (Ctp1), and Mre11-nuclease.PLoS Genet. 2013 Mar;9(3):e1003420. doi: 10.1371/journal.pgen.1003420. Epub 2013 Mar 28. PLoS Genet. 2013. PMID: 23555316 Free PMC article.
-
The Shu complex promotes error-free tolerance of alkylation-induced base excision repair products.Nucleic Acids Res. 2016 Sep 30;44(17):8199-215. doi: 10.1093/nar/gkw535. Epub 2016 Jun 13. Nucleic Acids Res. 2016. PMID: 27298254 Free PMC article.
-
Global Analysis of Furfural-Induced Genomic Instability Using a Yeast Model.Appl Environ Microbiol. 2019 Aug 29;85(18):e01237-19. doi: 10.1128/AEM.01237-19. Print 2019 Sep 15. Appl Environ Microbiol. 2019. PMID: 31300396 Free PMC article.
References
-
- Boiteux S, Guillet M. Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA Repair (Amst) 2004;3:1–12. - PubMed
-
- Lindahl T. Suppression of spontaneous mutagenesis in human cells by DNA base excision-repair. Mutat. Res. 2000;462:129–135. - PubMed
-
- Scharer OD, Jiricny J. Recent progress in the biology, chemistry and structural biology of DNA glycosylases. Bioessays. 2001;23:270–281. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

