Snu56p is required for Mer1p-activated meiotic splicing
- PMID: 18268012
- PMCID: PMC2293118
- DOI: 10.1128/MCB.00405-07
Snu56p is required for Mer1p-activated meiotic splicing
Abstract
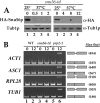
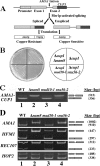
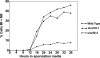
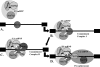
Alternative or regulated splicing can be applied to genes that are transcribed but whose products may be deleterious or unnecessary to the cell. In the yeast Saccharomyces cerevisiae, positive splicing regulation occurs during meiosis in which diploid cells divide to form haploid gametes. The Mer1 protein recruits the U1 snRNP to specific pre-mRNAs, permitting spliceosomal assembly and splicing. The mature transcripts are required for meiotic progression and, subsequently, sporulation. We have identified a novel allele (snu56-2) of the essential U1 snRNP protein Snu56p that exhibits a sporulation defect. Using a CUP1 reporter assay and reverse transcriptase PCR, we demonstrate that this allele specifically impairs Mer1p-activated splicing. This is not a reflection of a generally deficient spliceosome, as these cells splice vegetative transcripts efficiently. Furthermore, Snu56p depletion in vivo does not significantly impact mitotic splicing. Thus, its splicing function appears to be limited to Mer1p-activated meiosis-specific splicing. Two-hybrid studies indicate that Snu56p interacts with the other two U1 snRNP factors (Mer1p and Nam8p) required for this process. Interestingly, these two proteins do not interact, suggesting that Snu56p links pre-mRNA-bound Mer1p to Nam8p in the U1 snRNP. This work demonstrates that the Snu56 protein is required for splicing only during meiosis.
Figures



References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, NY.
-
- Baudin-Baillieu, A., E. Guillemet, C. Cullin, and F. Lacroute. 1997. Construction of a yeast strain deleted for the TRP1 promoter and coding region that enhances the efficiency of the polymerase chain reaction-disruption method. Yeast 13353-356. - PubMed
-
- Brow, D. A. 2002. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 36333-360. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
