Genome evolution in cyanobacteria: the stable core and the variable shell
- PMID: 18268351
- PMCID: PMC2268167
- DOI: 10.1073/pnas.0711165105
Genome evolution in cyanobacteria: the stable core and the variable shell
Abstract
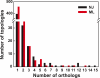
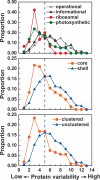
Cyanobacteria are the only known prokaryotes capable of oxygenic photosynthesis, the evolution of which transformed the biology and geochemistry of Earth. The rapid increase in published genomic sequences of cyanobacteria provides the first opportunity to reconstruct events in the evolution of oxygenic photosynthesis on the scale of entire genomes. Here, we demonstrate the overall phylogenetic incongruence among 682 orthologous protein families from 13 genomes of cyanobacteria. However, using principal coordinates analysis, we discovered a core set of 323 genes with similar evolutionary trajectories. The core set is highly conserved in amino acid sequence and contains genes encoding the major components in the photosynthetic and ribosomal apparatus. Many of the key proteins are encoded by genome-wide conserved small gene clusters, which often are indicative of protein-protein, protein-prosthetic group, and protein-lipid interactions. We propose that the macromolecular interactions in complex protein structures and metabolic pathways retard the tempo of evolution of the core genes and hence exert a selection pressure that restricts piecemeal horizontal gene transfer of components of the core. Identification of the core establishes a foundation for reconstructing robust organismal phylogeny in genome space. Our phylogenetic trees constructed from 16S rRNA gene sequences, concatenated orthologous proteins, and the core gene set all suggest that the ancestral cyanobacterium did not fix nitrogen and probably was a thermophilic organism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Bekker A, et al. Dating the rise of atmospheric oxygen. Nature. 2004;427:117–120. - PubMed
-
- Anbar AD, et al. A whiff of oxygen before the great oxidation event? Science. 2007;317:1903–1906. - PubMed
-
- Kaufman AJ, et al. Late Archean biospheric oxygenation and atmospheric evolution. Science. 2007;317:1900–1903. - PubMed
-
- Knoll AH, Summons RE, Waldbauer JR, Zumberge JE. In: Evolution of Primary Producers in the Sea. Falkowski PG, Knoll AH, editors. New York: Academic; 2007. pp. 133–163.
-
- Blankenship RE, Hartman H. The origin and evolution of oxygenic photosynthesis. Trends Biochem Sci. 1998;23:94–97. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

