Structure-function relationships among RNA-dependent RNA polymerases
- PMID: 18268843
- PMCID: PMC2441838
- DOI: 10.1007/978-3-540-75157-1_7
Structure-function relationships among RNA-dependent RNA polymerases
Abstract
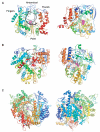
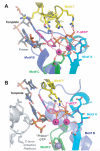
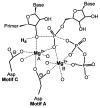
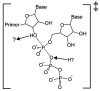
RNA-dependent RNA polymerases (RdRPs) play key roles in viral transcription and genome replication, as well as epigenetic and post-transcriptional control of cellular gene expression. In this article, we review the crystallographic, biochemical, and molecular genetic data available for viral RdRPs that have led to a detailed description of substrate and cofactor binding, fidelity of nucleotide selection and incorporation, and catalysis. It is likely that the cellular RdRPs will share some of the basic structural and mechanistic principles gleaned from studies of viral RdRPs. Therefore, studies of the viral RdRP establish a framework for the study of cellular RdRPs, an important yet understudied class of nucleic acid polymerases.
Figures



References
-
- Ago H, Adachi T, Yoshida A, Yamamoto M, Habuka N, Yatsunami K, Miyano M. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Structure Fold Des. 1999;7:1417–1426. - PubMed
-
- Andino R, Rieckhof GE, Baltimore D. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell. 1990;63:369–380. - PubMed
-
- Andino R, Boddeker N, Silvera D, Gamarnik AV. Intracellular determinants of picornavirus replication. Trends Microbiol. 1999;7:76–82. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

