Altered neuronal gene expression in brain regions differentially affected by Alzheimer's disease: a reference data set
- PMID: 18270320
- PMCID: PMC2826117
- DOI: 10.1152/physiolgenomics.00242.2007
Altered neuronal gene expression in brain regions differentially affected by Alzheimer's disease: a reference data set
Abstract
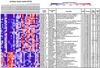
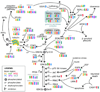
Alzheimer's Disease (AD) is the most widespread form of dementia during the later stages of life. If improved therapeutics are not developed, the prevalence of AD will drastically increase in the coming years as the world's population ages. By identifying differences in neuronal gene expression profiles between healthy elderly persons and individuals diagnosed with AD, we may be able to better understand the molecular mechanisms that drive AD pathogenesis, including the formation of amyloid plaques and neurofibrillary tangles. In this study, we expression profiled histopathologically normal cortical neurons collected with laser capture microdissection (LCM) from six anatomically and functionally discrete postmortem brain regions in 34 AD-afflicted individuals, using Affymetrix Human Genome U133 Plus 2.0 microarrays. These regions include the entorhinal cortex, hippocampus, middle temporal gyrus, posterior cingulate cortex, superior frontal gyrus, and primary visual cortex. This study is predicated on previous parallel research on the postmortem brains of the same six regions in 14 healthy elderly individuals, for which LCM neurons were similarly processed for expression analysis. We identified significant regional differential expression in AD brains compared with control brains including expression changes of genes previously implicated in AD pathogenesis, particularly with regard to tangle and plaque formation. Pinpointing the expression of factors that may play a role in AD pathogenesis provides a foundation for future identification of new targets for improved AD therapeutics. We provide this carefully phenotyped, laser capture microdissected intraindividual brain region expression data set to the community as a public resource.
Figures






References
-
- Alvarez A, Toro R, Caceres A, Maccioni RB. Inhibition of tau phosphorylating protein kinase cdk5 prevents beta-amyloid-induced neuronal death. FEBS Lett. 1999;459:421–426. - PubMed
-
- Barrachina M, Castano E, Ferrer I. TaqMan PCR assay in the control of RNA normalization in human post-mortem brain tissue. Neurochem Int. 2006;49:276–284. - PubMed
-
- Bhat RV, Budd Haeberlein SL, Avila J. Glycogen synthase kinase 3: a drug target for CNS therapies. J Neurochem. 2004;89:1313–1317. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30-AG-19610/AG/NIA NIH HHS/United States
- 1-R01-AG-023193/AG/NIA NIH HHS/United States
- P30 AG019610/AG/NIA NIH HHS/United States
- P50-AG-05681/AG/NIA NIH HHS/United States
- R01 AG023193/AG/NIA NIH HHS/United States
- K01-AG-024079/AG/NIA NIH HHS/United States
- P50 AG005128/AG/NIA NIH HHS/United States
- AG-05128/AG/NIA NIH HHS/United States
- U24 NS051872/NS/NINDS NIH HHS/United States
- K01 AG024079/AG/NIA NIH HHS/United States
- U01AG016976/AG/NIA NIH HHS/United States
- P01-AG-03991/AG/NIA NIH HHS/United States
- AG-O7367/AG/NIA NIH HHS/United States
- U01 AG016976/AG/NIA NIH HHS/United States
- P01 AG003991/AG/NIA NIH HHS/United States
- P50 AG005681/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

