Plasticity of the systemic inflammatory response to acute infection during critical illness: development of the riboleukogram
- PMID: 18270561
- PMCID: PMC2215774
- DOI: 10.1371/journal.pone.0001564
Plasticity of the systemic inflammatory response to acute infection during critical illness: development of the riboleukogram
Abstract
Background: Diagnosis of acute infection in the critically ill remains a challenge. We hypothesized that circulating leukocyte transcriptional profiles can be used to monitor the host response to and recovery from infection complicating critical illness.
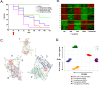
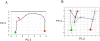
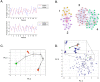
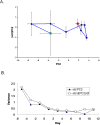
Methodology/principal findings: A translational research approach was employed. Fifteen mice underwent intratracheal injections of live P. aeruginosa, P. aeruginosa endotoxin, live S. pneumoniae, or normal saline. At 24 hours after injury, GeneChip microarray analysis of circulating buffy coat RNA identified 219 genes that distinguished between the pulmonary insults and differences in 7-day mortality. Similarly, buffy coat microarray expression profiles were generated from 27 mechanically ventilated patients every two days for up to three weeks. Significant heterogeneity of VAP microarray profiles was observed secondary to patient ethnicity, age, and gender, yet 85 genes were identified with consistent changes in abundance during the seven days bracketing the diagnosis of VAP. Principal components analysis of these 85 genes appeared to differentiate between the responses of subjects who did versus those who did not develop VAP, as defined by a general trajectory (riboleukogram) for the onset and resolution of VAP. As patients recovered from critical illness complicated by acute infection, the riboleukograms converged, consistent with an immune attractor.
Conclusions/significance: Here we present the culmination of a mouse pneumonia study, demonstrating for the first time that disease trajectories derived from microarray expression profiles can be used to quantitatively track the clinical course of acute disease and identify a state of immune recovery. These data suggest that the onset of an infection-specific transcriptional program may precede the clinical diagnosis of pneumonia in patients. Moreover, riboleukograms may help explain variance in the host response due to differences in ethnic background, gender, and pathogen. Prospective clinical trials are indicated to validate our results and test the clinical utility of riboleukograms.
Conflict of interest statement
Figures

References
-
- Smith JW, Gamelli RL, Jones SB, Shankar R. Immunologic responses to critical injury and sepsis. J Intensive Care Med. 2006;21:160–172. - PubMed
-
- Hotchkiss RS, Karl IE. The pathophysiology and treatment of sepsis. N Engl J Med. 2003;348:138–150. - PubMed
-
- Cobb JP, O'Keefe GE. Injury research in the genomic era. Lancet. 2004;363:2076–2083. - PubMed
-
- Osuchowski MF, Welch K, Siddiqui J, Remick DG. Circulating cytokine/inhibitor profiles reshape the understanding of the SIRS/CARS continuum in sepsis and predict mortality. J Immunol. 2006;177:1967–1974. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

