Targeted glycoproteomic identification of biomarkers for human breast carcinoma
- PMID: 18271524
- PMCID: PMC4932838
- DOI: 10.1021/pr700792g
Targeted glycoproteomic identification of biomarkers for human breast carcinoma
Abstract
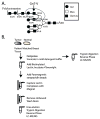
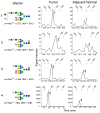
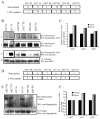
Glycosylation is a dynamic post-translational modification that changes during the development and progression of various malignancies. During the oncogenesis of breast carcinoma, the glycosyltransferase known as N-acetylglucosaminyltransferase Va (GnT-Va) transcript levels and activity are increased due to activated oncogenic signaling pathways. Elevated GnT-V levels leads to increased beta(1,6)-branched N-linked glycan structures on glycoproteins that can be measured using a specific carbohydrate binding protein or lectin known as L-PHA. L-PHA does not bind to nondiseased breast epithelial cells, but during the progression to invasive carcinoma, cells show a progressive increase in L-PHA binding. We have developed a procedure for intact protein L-PHA-affinity enrichment, followed by nanospray ionization mass spectrometry (NSI-MS/MS), to identify potential biomarkers for breast carcinoma. We identified L-PHA reactive glycoproteins from matched normal (nondiseased) and malignant tissue isolated from patients with invasive ductal breast carcinoma. Comparison analysis of the data identified 34 proteins that were enriched by L-PHA fractionation in tumor relative to normal tissue for at least 2 cases of ductal invasive breast carcinoma. Of these 34 L-PHA tumor enriched proteins, 12 are common to all 4 matched cases analyzed. These results indicate that lectin enrichment strategies targeting a particular glycan change associated with malignancy can be an effective method of identifying potential biomarkers for breast carcinoma.
Figures


References
-
- Jemal A, Murray T, Ward E, Samuels A, Tiwari RC, Ghafoor A, Feuer EJ, Thun MJ. Cancer statistics, 2005. CA Cancer J Clin. 2005;55(1):10–30. - PubMed
-
- Ries L, Eisner M, Kosary CL. SEER Cancer Statistics Review. National Cancer Institute; Bethesda, MD: 2005.
-
- Pucci-Minafra I, Cancemi P, Fontana S, Minafra L, Feo S, Becchi M, Freyria AM, Minafra S. Expanding the protein catalogue in the proteome reference map of human breast cancer cells. Proteomics. 2006;6(8):2609–2625. - PubMed
-
- Celis JE, Gromov P, Cabezon T, Moreira JM, Ambartsumian N, Sandelin K, Rank F, Gromova I. Proteomic characterization of the interstitial fluid perfusing the breast tumor microenvironment: a novel resource for biomarker and therapeutic target discovery. Mol Cell Proteomics. 2004;3(4):327–344. - PubMed
-
- Hudelist G, Singer CF, Pischinger KI, Kaserer K, Manavi M, Kubista E, Czerwenka KF. Proteomic analysis in human breast cancer: identification of a characteristic protein expression profile of malignant breast epithelium. Proteomics. 2006;6(6):1989–2002. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

