An erythroid differentiation signature predicts response to lenalidomide in myelodysplastic syndrome
- PMID: 18271621
- PMCID: PMC2235894
- DOI: 10.1371/journal.pmed.0050035
An erythroid differentiation signature predicts response to lenalidomide in myelodysplastic syndrome
Abstract
Background: Lenalidomide is an effective new agent for the treatment of patients with myelodysplastic syndrome (MDS), an acquired hematopoietic disorder characterized by ineffective blood cell production and a predisposition to the development of leukemia. Patients with an interstitial deletion of Chromosome 5q have a high rate of response to lenalidomide, but most MDS patients lack this deletion. Approximately 25% of patients without 5q deletions also benefit from lenalidomide therapy, but response in these patients cannot be predicted by any currently available diagnostic assays. The aim of this study was to develop a method to predict lenalidomide response in order to avoid unnecessary toxicity in patients unlikely to benefit from treatment.
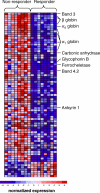
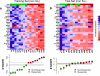
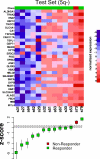
Methods and findings: Using gene expression profiling, we identified a molecular signature that predicts lenalidomide response. The signature was defined in a set of 16 pretreatment bone marrow aspirates from MDS patients without 5q deletions, and validated in an independent set of 26 samples. The response signature consisted of a cohesive set of erythroid-specific genes with decreased expression in responders, suggesting that a defect in erythroid differentiation underlies lenalidomide response. Consistent with this observation, treatment with lenalidomide promoted erythroid differentiation of primary hematopoietic progenitor cells grown in vitro.
Conclusions: These studies indicate that lenalidomide-responsive patients have a defect in erythroid differentiation, and suggest a strategy for a clinical test to predict patients most likely to respond to the drug. The experiments further suggest that the efficacy of lenalidomide, whose mechanism of action in MDS is unknown, may be due to its ability to induce erythroid differentiation.
Conflict of interest statement
Figures



Comment in
-
Solving the mystery of myelodysplasia.PLoS Med. 2008 Feb;5(2):e40. doi: 10.1371/journal.pmed.0050040. PLoS Med. 2008. PMID: 18271622 Free PMC article. Review.
References
-
- Deangelo D, Stone R, editors. Myelodysplastic syndromes: biology and treatment. Philadelphia: Elsevier; 2005. pp. 1195–1208.
-
- Heaney ML, Golde DW. Myelodysplasia. N Engl J Med. 1999;340:1649–1660. - PubMed
-
- List A, Dewald G, Bennett J, Giagounidis A, Raza A, et al. Lenalidomide in the myelodysplastic syndrome with chromosome 5q deletion. N Engl J Med. 2006;355:1456–1465. - PubMed
-
- List AF, Kurtin S, Roe D, Buresh A, Mahadevan D, et al. Efficacy of lenalidomide in myelodysplastic syndromes. N Engl J Med. 2005;352:549–557. - PubMed
-
- Raza A, Reeves JA, Feldman EJ, Dewald GW, Bennett JM, et al. Phase II study of lenalidomide in transfusion-dependent, low- and intermediate-1-risk myelodysplastic syndromes with karyotypes other than deletion 5q. Blood. 2008;11:86–93. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

