Still looking for the magic spot: the crystallographically defined binding site for ppGpp on RNA polymerase is unlikely to be responsible for rRNA transcription regulation
- PMID: 18272182
- PMCID: PMC2317782
- DOI: 10.1016/j.jmb.2008.01.042
Still looking for the magic spot: the crystallographically defined binding site for ppGpp on RNA polymerase is unlikely to be responsible for rRNA transcription regulation
Erratum in
- J Mol Biol. 2008 Jun 20;379(5):1130
Abstract
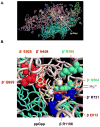
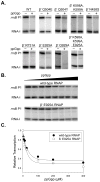
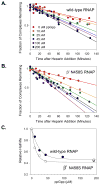
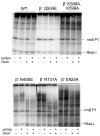
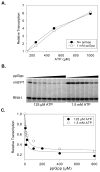
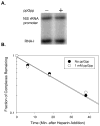
Identification of the RNA polymerase (RNAP) binding site for ppGpp, a central regulator of bacterial transcription, is crucial for understanding its mechanism of action. A recent high-resolution X-ray structure defined a ppGpp binding site on Thermus thermophilus RNAP. We report here effects of ppGpp on 10 mutant Escherichia coli RNAPs with substitutions for the analogous residues within 3-4 A of the ppGpp binding site in the T. thermophilus cocrystal. None of the substitutions in E. coli RNAP significantly weakened its responses to ppGpp. This result differs from the originally reported finding of a substitution in E. coli RNAP eliminating ppGpp function. The E. coli RNAPs used in that study likely lacked stoichiometric amounts of omega, an RNAP subunit required for responses of RNAP to ppGpp, in part explaining the discrepancy. Furthermore, we found that ppGpp did not inhibit transcription initiation by T. thermophilus RNAP in vitro or shorten the lifetimes of promoter complexes containing T. thermophilus RNAP, in contrast to the conclusion in the original report. Our results suggest that the ppGpp binding pocket identified in the cocrystal is not the one responsible for regulation of E. coli ribosomal RNA transcription initiation and highlight the importance of inclusion of omega in bacterial RNAP preparations.
Figures
References
-
- Cashel M, Gentry DR, Hernandez VH, Vinella D. The Stringent Response. In: Neidhardt FC, editor. Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press; Washington, D.C: 1996. pp. 1458–1496.
-
- Murray HD, Schneider DA, Gourse RL. Control of rRNA expression by small molecules is dynamic and nonredundant. Mol Cell. 2003;12:125–134. - PubMed
-
- Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, Foster JW, Gourse RL. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell. 2004;118:311–322. - PubMed
-
- Bernardo LM, Johansson LU, Solera D, Skarfstad E, Shingler V. The guanosine tetraphosphate (ppGpp) alarmone, DksA and promoter affinity for RNA polymerase in regulation of sigma-dependent transcription. Mol Microbiol. 2006;60:749–764. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

