Insights into bunyavirus architecture from electron cryotomography of Uukuniemi virus
- PMID: 18272496
- PMCID: PMC2268144
- DOI: 10.1073/pnas.0708738105
Insights into bunyavirus architecture from electron cryotomography of Uukuniemi virus
Abstract
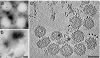
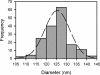
Bunyaviridae is a large family of viruses that have gained attention as "emerging viruses" because many members cause serious disease in humans, with an increasing number of outbreaks. These negative-strand RNA viruses possess a membrane envelope covered by glycoproteins. The virions are pleiomorphic and thus have not been amenable to structural characterization using common techniques that involve averaging of electron microscopic images. Here, we determined the three-dimensional structure of a member of the Bunyaviridae family by using electron cryotomography. The genome, incorporated as a complex with the nucleoprotein inside the virions, was seen as a thread-like structure partially interacting with the viral membrane. Although no ordered nucleocapsid was observed, lateral interactions between the two membrane glycoproteins determine the structure of the viral particles. In the most regular particles, the glycoprotein protrusions, or "spikes," were seen to be arranged on an icosahedral lattice, with T = 12 triangulation. This arrangement has not yet been proven for a virus. Two distinctly different spike conformations were observed, which were shown to depend on pH. This finding is reminiscent of the fusion proteins of alpha-, flavi-, and influenza viruses, in which conformational changes occur in the low pH of the endosome to facilitate fusion of the viral and host membrane during viral entry.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Calisher CH. History, classification, and taxonomy of viruses in the family Bunyaviridae. In: Elliott RM, editor. The Bunyaviridae. New York: Plenum; 1996. pp. 1–17.
-
- Soldan SS, Gonzalez-Scarano F. Emerging infectious diseases: The Bunyaviridae. J Neurovirol. 2005;11:412–423. - PubMed
-
- Schmaljohn CS, Nichol ST. Bunyaviridae. In: Knipe DM, Howley PM, editors. Fields Virology. 5th Ed. Vol 2. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 1741–1789.
-
- Hacker JK, Hardy JL. Adsorptive endocytosis of California encephalitis virus into mosquito and mammalian cells: A role for G1. Virology. 1997;235:40–47. - PubMed
-
- Jin M, et al. Hantaan virus enters cells by clathrin-dependent receptor-mediated endocytosis. Virology. 2002;294:60–69. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

