Functional characterization of Rab7 mutant proteins associated with Charcot-Marie-Tooth type 2B disease
- PMID: 18272684
- PMCID: PMC6671532
- DOI: 10.1523/JNEUROSCI.3677-07.2008
Functional characterization of Rab7 mutant proteins associated with Charcot-Marie-Tooth type 2B disease
Abstract
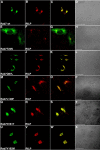
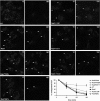
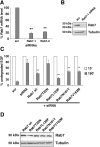
Charcot-Marie-Tooth (CMT) type 2 neuropathies are a group of autosomal-dominant axonal disorders genetically and clinically heterogeneous. In particular, CMT type 2B (CMT2B) neuropathies are characterized by severe sensory loss, often complicated by infections, arthropathy, and amputations. Recently, four missense mutations in the small GTPase Rab7 associated with the Charcot-Marie Tooth type 2B phenotype have been identified. These mutations target highly conserved amino acid residues. However, nothing is known about whether and how these mutations affect Rab7 function. We investigated the biochemical and functional properties of three of the mutant proteins. Interestingly, all three proteins exhibited higher nucleotide exchange rates and hydrolyzed GTP slower than the wild-type protein. In addition, whereas 23% of overexpressed wild-type Rab7 was GTP bound in HeLa cells, the large majority of the mutant proteins (82-89%) were in the GTP-bound form, consistent with the data on GTP hydrolysis and exchange rates. The CMT2B-associated Rab7 proteins were also able to bind the Rab7 effector RILP (Rab-interacting lysosomal protein) and to rescue Rab7 function after silencing. Altogether, these data demonstrate that all tested CMT2B-associated Rab7 mutations are mechanistically similar, suggesting that activated forms of the Rab7 are responsible for CMT2B disease.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
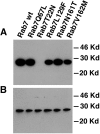
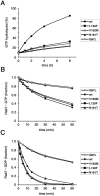
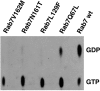
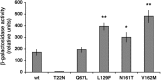


References
-
- Auer-Grumbach M, Mauko B, Auer-Grumbach P, Pieber TR. Molecular genetics of hereditary sensory neuropathies. Neuromolecular Med. 2006;8:147–158. - PubMed
-
- Bartel P, Chien CT, Sternglanz R, Fields S. Elimination of false positives that arise in using the two-hybrid system. Biotechniques. 1993;14:920–924. - PubMed
-
- Bronfman FC, Escudero CA, Weis J, Kruttgen A. Endosomal transport of neurotrophins: roles in signaling and neurodegenerative diseases. Dev Neurobiol. 2007;67:1183–1203. - PubMed
-
- Bucci C, Chiariello M. Signal transduction gRABs attention. Cell Signal. 2006;18:1–8. - PubMed
-
- Bucci C, Parton RG, Mather IH, Stunnenberg H, Simons K, Hoflack B, Zerial M. The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell. 1992;70:715–728. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
