Salmonella enterica serovar Senftenberg human clinical isolates lacking SPI-1
- PMID: 18272702
- PMCID: PMC2292908
- DOI: 10.1128/JCM.01255-07
Salmonella enterica serovar Senftenberg human clinical isolates lacking SPI-1
Abstract
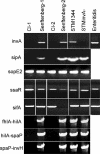
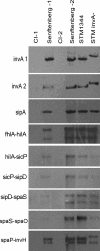
Nontyphoidal Salmonella species cause gastrointestinal disease worldwide. The prevailing theory of Salmonella enteropathogenesis is that bacterial invasion of the intestinal epithelium is essential for virulence and that this requires the virulence-associated genomic region Salmonella pathogenicity island 1 (SPI-1). Recent studies of Salmonella enterica infection models have demonstrated that enterocolitis and diarrhea in mice and cows can occur independently of SPI-1. In this study, we sought to confirm whether two S. enterica serovar Senftenberg clinical isolates lacked genes essential for SPI-1 function. Two clinical strains were isolated and identified as being S. enterica serovar Senftenberg from four stool samples from a food-borne disease outbreak affecting seven individuals in Shenzhen, Guangdong Province, China, using conventional methods, pulsed-field gel electrophoresis and multilocus sequence typing. The possibility of coinfection with other potential bacteria or usual viruses was excluded. Two isolates were analyzed for the presence of invA, sipA, ssaR, sifA, and sopE2 by PCR and Southern blotting and were then assayed for the presence of SPI-1 by PCR and long-range PCR for fhlA-hilA, hilA-spaP, and spaP-invH and Southern blot analysis. A long-range PCR fragment from fhlA to mutS covering the 5' and 3' flanks of SPI-1 was also amplified from the two clinical isolates and sequenced. In addition, the two clinical isolates were assayed for enteroinvasiveness in vitro. Murine infection models were also examined. Biochemical tests and serotyping confirmed that the two clinical isolates are S. enterica serovar Senftenberg. However, they lacked genes critical for SPI-1 function but contained SPI-2 genes and were attenuated for the invasion of cultured intestinal epithelial cells. In conclusion, clinical S. enterica serovar Senftenberg strains isolated from a food-borne disease outbreak lack the invasion-associated locus SPI-1, indicating that SPI-1 is not essential for human gastroenteritis.
Figures


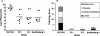
References
-
- Arnold, T., H. C. Scholz, H. Marg, U. Rosler, and A. Hensel. 2004. Impact of invA-PCR and culture detection methods on occurrence and survival of Salmonella in the flesh, internal organs and lymphoid tissues of experimentally infected pigs. J. Vet. Med. B Infect. Dis. Vet. Public Health 51459-463. - PubMed
-
- Barthel, M., S. Hapfelmeier, L. Quintanilla-Martinez, M. Kremer, M. Rohde, M. Hogardt, K. Pfeffer, H. Russmann, and W. D. Hardt. 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect. Immun. 712839-2858. - PMC - PubMed
-
- Bulte, M., and P. Jakob. 1995. The use of a PCR-generated invA probe for the detection of Salmonella spp. in artificially and naturally contaminated foods. Int. J. Food Microbiol. 26335-344. - PubMed
-
- Cocolin, L., M. Manzano, G. Astori, G. A. Botta, C. Cantoni, and G. Comi. 1998. A highly sensitive and fast non-radioactive method for the detection of polymerase chain reaction products from Salmonella serovars, such as Salmonella Typhi, in blood specimens. FEMS Immunol. Med. Microbiol. 22233-239. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

