A novel switch region regulates H-ras membrane orientation and signal output
- PMID: 18273062
- PMCID: PMC2265749
- DOI: 10.1038/emboj.2008.10
A novel switch region regulates H-ras membrane orientation and signal output
Abstract
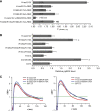
The plasma membrane nanoscale distribution of H-ras is regulated by guanine nucleotide binding. To explore the structural basis of H-ras membrane organization, we combined molecular dynamic simulations and medium-throughput FRET measurements on live cells. We extracted a set of FRET values, termed a FRET vector, to describe the lateral segregation and orientation of H-ras with respect to a large set of nanodomain markers. We show that mutation of basic residues in helix alpha4 or the hypervariable region (HVR) selectively alter the FRET vectors of GTP- or GDP-loaded H-ras, demonstrating a critical role for these residues in stabilizing GTP- or GDP-H-ras interactions with the plasma membrane. By a similar analysis, we find that the beta2-beta3 loop and helix alpha5 are involved in a novel conformational switch that operates through helix alpha4 and the HVR to reorient the H-ras G-domain with respect to the plasma membrane. Perturbation of these switch elements enhances MAPK activation by stabilizing GTP-H-ras in a more productive signalling conformation. The results illustrate how the plasma membrane spatially constrains signalling conformations by acting as a semi-neutral interaction partner.
Figures



References
-
- Abankwa D, Vogel H (2007) A FRET map of membrane anchors suggests distinct microdomains of heterotrimeric G proteins. J Cell Sci 120: 2953–2962 - PubMed
-
- Diggle PJ, Lange N, Benes FM (1991) Analysis of variance for replicated spatial point patterns in clinical neuroanatomy. J Am Stat Assoc 86: 618–625
-
- Diggle PJ, Mateu J, Clough HE (2000) A comparison between parametric and non-parametric approaches to the analysis of replicated spatial point patterns. Adv Appl Probab 32: 331–343
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

