Functional protein divergence in the evolution of Homo sapiens
- PMID: 18279504
- PMCID: PMC2374701
- DOI: 10.1186/gb-2008-9-2-r33
Functional protein divergence in the evolution of Homo sapiens
Abstract
Background: Protein-coding regions in a genome evolve by sequence divergence and gene gain and loss, altering the gene content of the organism. However, it is not well understood how this has given rise to the enormous diversity of metazoa present today.
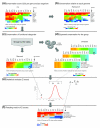
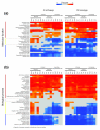
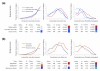
Results: To obtain a global view of human genomic evolution, we quantify the divergence of proteins by functional category at different evolutionary distances from human.
Conclusion: This analysis highlights some general systems-level characteristics of human evolution: regulatory processes, such as signal transducers, transcription factors and receptors, have a high degree of plasticity, while core processes, such as metabolism, transport and protein synthesis, are largely conserved. Additionally, this study reveals a dynamic picture of selective forces at short, medium and long evolutionary timescales. Certain functional categories, such as 'development' and 'organogenesis', exhibit temporal patterns of sequence divergence in eukaryotes relative to human. This framework for a grammar of human evolution supports previously postulated theories of robustness and evolvability.
Figures


References
-
- Dayhoff M, Schwartz R, Orcutt B. A model of evolutionary change in proteins. In: Dayhoff M, editor. Atlas of Protein Sequence and Structure. Vol. 5. Silver Springs, MD: National Biomedical Research Foundation; 1978. pp. 345–352.
-
- Hillier LW, Miller W, Birney E, Warren W, Hardison RC, Ponting CP, Bork P, Burt DW, Groenen MA, Delany ME, Dodgson JB, Chinwalla AT, Cliften PF, Clifton SW, Delehaunty KD, Fronick C, Fulton RS, Graves TA, Kremitzki C, Layman D, Magrini V, McPherson JD, Miner TL, Minx P, Nash WE, Nhan MN, Nelson JO, Oddy LG, Pohl CS, Randall-Maher J, et al. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

