Mouse N-acetyltransferase type 2, the homologue of human N-acetyltransferase type 1
- PMID: 18280460
- PMCID: PMC2279149
- DOI: 10.1016/j.bcp.2007.12.012
Mouse N-acetyltransferase type 2, the homologue of human N-acetyltransferase type 1
Abstract
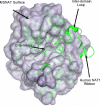
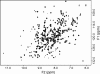
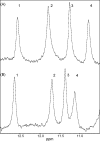
There is increasing evidence that human arylamine N-acetyltransferase type 1 (NAT1, EC 2.3.1.5), although first identified as a homologue of a drug-metabolising enzyme, appears to be a marker in human oestrogen receptor positive breast cancer. Mouse Nat2 is the mouse equivalent of human NAT1. The development of mouse models of breast cancer is important, and it is essential to explore the biological role of mouse Nat2. We have therefore produced mouse Nat2 as a recombinant protein and have investigated its substrate specificity profile in comparison with human NAT1. In addition, we have tested the effects of inhibitors on mouse Nat2, including compounds which are endogenous and exogenous steroids. We show that tamoxifen, genistein and diethylstilbestrol inhibit mouse Nat2. The steroid analogue, bisphenol A, also inhibits mouse Nat2 enzymic activity and is shown by NMR spectroscopy, through shifts in proton peaks, to bind close to the active site. A three-dimensional structure for human NAT1 has recently been released, and we have used this crystal structure to generate a model of the mouse Nat2 structure. We propose that a conformational change in the structure is required in order for ligands to bind to the active site of the protein.
Figures

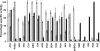



References
-
- Sim E., Westwood I., Fullam E. Arylamine N-acetyltransferases. Expert Opin. Drug Metab. Toxicol. 2007;3:169–184. - PubMed
-
- Kawamura A., Graham J., Mushtaq A., Tsiftsoglou S.A., Vath G.M., Hanna P.E. Eukaryotic arylamine N-acetyltransferase Investigation of substrate specificity by high-throughput screening. Biochem. Pharmacol. 2005;69:347–359. - PubMed
-
- Gruvberger S., Ringner M., Chen Y., Panavally S., Saal L.H., Borg A. Estrogen receptor status in breast cancer is associated with remarkably distinct gene expression patterns. Cancer Res. 2001;61:5979–5984. - PubMed
-
- Hess K.R., Anderson K., Symmans W.F., Valero V., Ibrahim N., Mejia J.A. Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J. Clin. Oncol. 2006;24:4236–4244. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

