SprB is a cell surface component of the Flavobacterium johnsoniae gliding motility machinery
- PMID: 18281397
- PMCID: PMC2293251
- DOI: 10.1128/JB.01904-07
SprB is a cell surface component of the Flavobacterium johnsoniae gliding motility machinery
Abstract
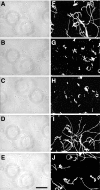
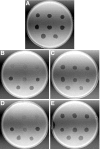
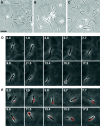
Cells of the gliding bacterium Flavobacterium johnsoniae move rapidly over surfaces by an unknown mechanism. Transposon insertions in sprB resulted in cells that were defective in gliding. SprB is a highly repetitive 669-kDa cell surface protein, and antibodies against SprB inhibited the motility of wild-type cells. Polystyrene microspheres coated with antibodies against SprB attached to and were rapidly propelled along the cell surface, suggesting that SprB is one of the outermost components of the motility machinery. The movement of SprB along the cell surface supports a model of gliding motility in which motors anchored to the cell wall rapidly propel cell surface adhesins.
Figures





References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. - PubMed
-
- Bardy, S. L., S. Y. M. Ng, and K. F. Jarrell. 2003. Prokaryotic motility structures. Microbiology 149295-304. - PubMed
-
- Beatson, P. J., and K. C. Marshall. 1994. A proposed helical mechanism for gliding motility in three gliding bacteria (order Cytophagales). Can. J. Microbiol. 40173-183.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

