Lgt: a family of cytotoxic glucosyltransferases produced by Legionella pneumophila
- PMID: 18281405
- PMCID: PMC2293231
- DOI: 10.1128/JB.01798-07
Lgt: a family of cytotoxic glucosyltransferases produced by Legionella pneumophila
Abstract
Legionella pneumophila is a facultative intracellular pathogen responsible for severe lung disease in humans, known as legionellosis or Legionnaires' disease. Previously, we reported on the approximately 60-kDa glucosyltransferase (Lgt1) from Legionella pneumophila, which modified eukaryotic elongation factor 1A. In the present study, using L. pneumophila Philadelphia-1, Lens, Paris, and Corby genome databases, we identified several genes coding for proteins with considerable sequence homology to Lgt1. These new enzymes form three subfamilies, termed Lgt1 to -3, glucosylate mammalian elongation factor eEF1A at serine-53, inhibit its activity, and subsequently kill target eukaryotic cells. Expression studies on L. pneumophila grown in broth medium or in Acanthamoeba castellanii revealed that production of Lgt1 was maximal at stationary phase of broth culture or during the late phase of Legionella-host cell interaction, respectively. In contrast, synthesis of Lgt3 peaked during the lag phase of liquid culture and at early steps of bacterium-amoeba interaction. Thus, the data indicate that members of the L. pneumophila glucosyltransferase family are differentially regulated, affect protein synthesis of host cells, and represent potential virulence factors of Legionella.
Figures






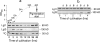
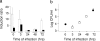
References
-
- Aktories, K., and J. T. Barbieri. 2005. Bacterial cytotoxins: targeting eukaryotic switches. Nat. Rev. Microbiol. 3397-410. - PubMed
-
- Bachman, M. A., and M. S. Swanson. 2001. RpoS co-operates with other factors to induce Legionella pneumophila virulence in the stationary phase. Mol. Microbiol. 401201-1214. - PubMed
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

