Syntrophorhabdus aromaticivorans gen. nov., sp. nov., the first cultured anaerobe capable of degrading phenol to acetate in obligate syntrophic associations with a hydrogenotrophic methanogen
- PMID: 18281436
- PMCID: PMC2292594
- DOI: 10.1128/AEM.02378-07
Syntrophorhabdus aromaticivorans gen. nov., sp. nov., the first cultured anaerobe capable of degrading phenol to acetate in obligate syntrophic associations with a hydrogenotrophic methanogen
Abstract
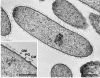
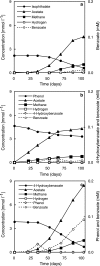
Phenol degradation under methanogenic conditions has long been studied, but the anaerobes responsible for the degradation reaction are still largely unknown. An anaerobe, designated strain UI(T), was isolated in a pure syntrophic culture. This isolate is the first tangible, obligately anaerobic, syntrophic substrate-degrading organism capable of oxidizing phenol in association with an H(2)-scavenging methanogen partner. Besides phenol, it could metabolize p-cresol, 4-hydroxybenzoate, isophthalate, and benzoate. During the degradation of phenol, a small amount of 4-hydroxybenzoate (a maximum of 4 microM) and benzoate (a maximum of 11 microM) were formed as transient intermediates. When 4-hydroxybenzoate was used as the substrate, phenol (maximum, 20 microM) and benzoate (maximum, 92 microM) were detected as intermediates, which were then further degraded to acetate and methane by the coculture. No substrates were found to support the fermentative growth of strain UI(T) in pure culture, although 88 different substrates were tested for growth. 16S rRNA gene sequence analysis indicated that strain UI(T) belongs to an uncultured clone cluster (group TA) at the family (or order) level in the class Deltaproteobacteria. Syntrophorhabdus aromaticivorans gen. nov., sp. nov., is proposed for strain UI(T), and the novel family Syntrophorhabdaceae fam. nov. is described. Peripheral 16S rRNA gene sequences in the databases indicated that the proposed new family Syntrophorhabdaceae is largely represented by abundant bacteria within anaerobic ecosystems mainly decomposing aromatic compounds.
Figures


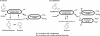
References
-
- Béchard, G., J.-G. Bisaillon, R. Beaudet, and M. Sylvestre. 1990. Degradation of phenol by a bacterial consortium under methanogenic conditions. Can. J. Microbiol. 36:573-578.
-
- Bisaillon, J. G., F. Lépine, and R. Beaudet. 1991. Study of the methanogenic degradation of phenol via carboxylation to benzoate. Can. J. Microbiol. 37:573-576.
-
- Bisaillon, J. G., F. Lépine, R. Beaudet, and M. Sylvestre. 1993. Potential for carboxylation-dehydroxylation of phenolic compounds by a methanogenic consortium. Can. J. Microbiol. 39:642-648. - PubMed
-
- Breitenstein, A., J. Wiegel, C. Haertig, N. Weiss, J. R. Andreesen, and U. Lechner. 2002. Reclassification of Clostridium hydroxybenzoicum as Sedimentibacter hydroxybenzoicus gen. nov., comb. nov., and description of Sedimentibacter saalensis sp. nov. Int. J. Syst. Evol. Microbiol. 52:801-807. - PubMed
-
- Cervantes, F. J., S. van der Velde, G. Lettinga, and J. A. Field. 2000. Quinones as terminal electron acceptors for anaerobic microbial oxidation of phenolic compounds. Biodegradation 11:313-321. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

