Allele-specific nuclear positioning of the monoallelically expressed astrocyte marker GFAP
- PMID: 18281462
- PMCID: PMC2238670
- DOI: 10.1101/gad.1634608
Allele-specific nuclear positioning of the monoallelically expressed astrocyte marker GFAP
Abstract
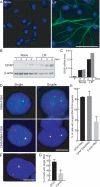
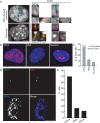
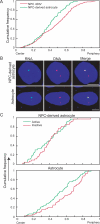
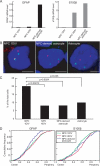
Chromosomes and genes are nonrandomly arranged within the mammalian cell nucleus. However, the functional significance of nuclear positioning in gene expression is unclear. Here we directly probed the relationship between nuclear positioning and gene activity by comparing the location of the active and inactive copies of a monoallelically expressed gene in single cell nuclei. We demonstrate that the astrocyte-specific marker GFAP (glial fibrillary acidic protein) is monoallelically expressed in cortical astrocytes. Selection of the active allele occurs in a stochastic manner and is generally maintained through cell division. Taking advantage of the monoallelic expression of GFAP, we show that the functionally distinct alleles occupy differential radial positions within the cell nucleus and differentially associate with intranuclear compartments. In addition, coordinately regulated astrocyte-specific genes on distinct chromosomes spatially associate in their inactive state and dissociate upon activation. These results provide direct evidence for function-related differential positioning of individual gene alleles within the interphase nucleus.
Figures

References
-
- Baxter J., Merkenschlager M., Fisher A.G. Nuclear organisation and gene expression. Curr. Opin. Cell Biol. 2002;14:372–376. - PubMed
-
- Bickmore W.A., Mahy N.L., Chambeyron S. Do higher-order chromatin structure and nuclear reorganization play a role in regulating Hox gene expression during development? Cold Spring Harb. Symp. Quant. Biol. 2004;69:251–257. - PubMed
-
- Castleman K.R. Digital image processing. Prentice Hall; Englewood Cliffs, NJ: 1996.
-
- Chakalova L., Debrand E., Mitchell J.A., Osborne C.S., Fraser P. Replication and transcription: Shaping the landscape of the genome. Nat. Rev. 2005;6:669–677. - PubMed
-
- Cremer T., Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2001;2:292–301. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
