CrossSearch, a user-friendly search engine for detecting chemically cross-linked peptides in conjugated proteins
- PMID: 18281724
- PMCID: PMC2401330
- DOI: 10.1074/mcp.M800020-MCP200
CrossSearch, a user-friendly search engine for detecting chemically cross-linked peptides in conjugated proteins
Abstract
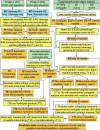
Chemical cross-linking and high resolution MS have been integrated successfully to capture protein interactions and provide low resolution structural data for proteins that are refractive to analyses by NMR or crystallography. Despite the versatility of these combined techniques, the array of products that is generated from the cross-linking and proteolytic digestion of proteins is immense and generally requires the use of labeling strategies and/or data base search algorithms to distinguish actual cross-linked peptides from the many side products of cross-linking. Most strategies reported to date have focused on the analysis of small cross-linked protein complexes (<60 kDa) because the number of potential forms of covalently modified peptides increases dramatically with the number of peptides generated from the digestion of such complexes. We report herein the development of a user-friendly search engine, CrossSearch, that provides the foundation for an overarching strategy to detect cross-linked peptides from the digests of large (>or=170-kDa) cross-linked proteins, i.e. conjugates. Our strategy combines the use of a low excess of cross-linker, data base searching, and Fourier transform ion cyclotron resonance MS to experimentally minimize and theoretically cull the side products of cross-linking. Using this strategy, the (alpha beta gamma delta)(4) phosphorylase kinase model complex was cross-linked to form with high specificity a 170-kDa betagamma conjugate in which we identified residues involved in the intramolecular cross-linking of the 125-kDa beta subunit between its regulatory N terminus and its C terminus. This finding provides an explanation for previously published homodimeric two-hybrid interactions of the beta subunit and suggests a dynamic structural role for the regulatory N terminus of that subunit. The results offer proof of concept for the CrossSearch strategy for analyzing conjugates and are the first to reveal a tertiary structural element of either homologous alpha or beta regulatory subunit of phosphorylase kinase.
Figures




References
-
- Nadeau, O. W., and Carlson, G. M. (2005) Protein interactions captured by chemical cross-linking, in Protein-Protein Interactions, A Molecular Cloning Manual (Golemis, E., and Adams, P. D., eds) 2nd Ed., pp. 105–127, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
-
- Sinz, A. (2006) Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein-protein interactions. Mass Spectrom. Rev. 25 663–682 - PubMed
-
- Nadeau, O. W. (2006) Protein interaction analysis: chemical cross-linking, in Encyclopedic Reference of Genomics and Proteomics in Molecular Medicine (Ganten, D., and Ruckpaul, K., eds) pp. 1506–1509, Springer, Berlin
-
- Soderblom, E. J., and Goshe, M. B. (2006) Collision-induced dissociative chemical cross-linking reagents and methodology: applications to protein structural characterization using tandem mass spectrometry analysis. Anal. Chem. 78 8059–8068 - PubMed
-
- Nadeau, O. W., and Carlson, G. M. (eds) (2002) Protein-protein interactions, in Protein-Protein Interactions, A Molecular cloning Manual (Golemis, E., ed) 1st Ed., pp. 75–91, Cold Spring Harbor Press, Cold Spring Harbor, NY
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

