Structural insight into epitopes in the pregnancy-associated malaria protein VAR2CSA
- PMID: 18282103
- PMCID: PMC2242842
- DOI: 10.1371/journal.ppat.0040042
Structural insight into epitopes in the pregnancy-associated malaria protein VAR2CSA
Abstract
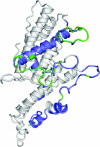
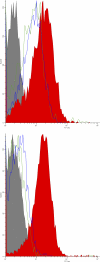
Pregnancy-associated malaria is caused by Plasmodium falciparum malaria parasites binding specifically to chondroitin sulfate A in the placenta. This sequestration of parasites is a major cause of low birth weight in infants and anemia in the mothers. VAR2CSA, a polymorphic multi-domain protein of the PfEMP1 family, is the main parasite ligand for CSA binding, and identification of protective antibody epitopes is essential for VAR2CSA vaccine development. Attempts to determine the crystallographic structures of VAR2CSA or its domains have not been successful yet. In this study, we propose 3D models for each of the VAR2CSA DBL domains and we show that regions in the fold of VAR2CSA inter-domain 2 and a PfEMP1 CIDR domain seem to be homologous to the EBA-175 and Pk alpha-DBL fold. This suggests that ID2 could be a functional domain. We also identify regions of VAR2CSA present on the surface of native VAR2CSA by comparing reactivity of plasma containing anti-VAR2CSA antibodies in peptide array experiments before and after incubation with native VAR2CSA. By this method we identify conserved VAR2CSA regions targeted by antibodies that react with the native molecule expressed on infected erythrocytes. By mapping the data onto the DBL models we present evidence suggesting that the S1+S2 DBL sub-domains are generally surface-exposed in most domains, whereas the S3 sub-domains are less exposed in native VAR2CSA. These results comprise an important step towards understanding the structure of VAR2CSA on the surface of CSA-binding infected erythrocytes.
Conflict of interest statement
Figures


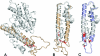



References
-
- Baruch DI, Pasloske BL, Singh HB, Bi X, Ma XC, et al. Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell. 1995;82:77–87. - PubMed
-
- Weiss L. The spleen in malaria: The role of barrier cells. Immunol Lett. 1990;25:165–172. - PubMed
-
- Kraemer SM, Smith JD. A family affair: var genes, PfEMP1 binding, and malaria disease. Curr Opin Microbiol. 2006;9:374–380. - PubMed
-
- Fried M, Duffy PE. Adherence of Plasmodium falciparum to chondroitin sulfate A in the human placenta. Science. 1996;272:1502–1504. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

