Epigenetic inheritance based evolution of antibiotic resistance in bacteria
- PMID: 18282299
- PMCID: PMC2262874
- DOI: 10.1186/1471-2148-8-52
Epigenetic inheritance based evolution of antibiotic resistance in bacteria
Abstract
Background: The evolution of antibiotic resistance in bacteria is a topic of major medical importance. Evolution is the result of natural selection acting on variant phenotypes. Both the rigid base sequence of DNA and the more plastic expression patterns of the genes present define phenotype.
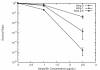
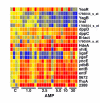
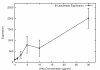
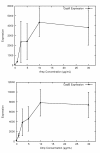
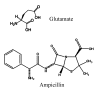
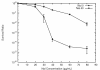
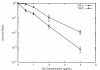
Results: We investigated the evolution of resistant E. coli when exposed to low concentrations of antibiotic. We show that within an isogenic population there are heritable variations in gene expression patterns, providing phenotypic diversity for antibiotic selection to act on. We studied resistance to three different antibiotics, ampicillin, tetracycline and nalidixic acid, which act by inhibiting cell wall synthesis, protein synthesis and DNA synthesis, respectively. In each case survival rates were too high to be accounted for by spontaneous DNA mutation. In addition, resistance levels could be ramped higher by successive exposures to increasing antibiotic concentrations. Furthermore, reversion rates to antibiotic sensitivity were extremely high, generally over 50%, consistent with an epigenetic inheritance mode of resistance. The gene expression patterns of the antibiotic resistant E. coli were characterized with microarrays. Candidate genes, whose altered expression might confer survival, were tested by driving constitutive overexpression and determining antibiotic resistance. Three categories of resistance genes were identified. The endogenous beta-lactamase gene represented a cryptic gene, normally inactive, but when by chance expressed capable of providing potent ampicillin resistance. The glutamate decarboxylase gene, in contrast, is normally expressed, but when overexpressed has the incidental capacity to give an increase in ampicillin resistance. And the DAM methylase gene is capable of regulating the expression of other genes, including multidrug efflux pumps.
Conclusion: In this report we describe the evolution of antibiotic resistance in bacteria mediated by the epigenetic inheritance of variant gene expression patterns. This provides proof in principle that epigenetic inheritance, as well as DNA mutation, can drive evolution.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

