Conserved themes in small-RNA-mediated transposon control
- PMID: 18282709
- PMCID: PMC2995447
- DOI: 10.1016/j.tcb.2008.01.004
Conserved themes in small-RNA-mediated transposon control
Abstract
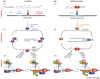
Eukaryotes are engaged in a constant struggle against transposable elements, which have invaded and profoundly shaped their genomes. Over the past decade, a growing body of evidence has pointed to a role for small RNAs in transposon defense. Although the strategies used in different organisms vary in their details, they have strikingly similar general properties. Basically, all mechanisms consist of three components. First, transposon detection prompts the production of small RNAs, which are Piwi-interacting RNAs in some organisms and small interfering RNAs in others. Second, the population of small RNAs targeting active transposons is amplified through an RNA-dependent RNA polymerase-based or Slicer-based mechanism. Third, small RNAs are incorporated into Argonaute- or Piwi-containing effector complexes, which target transposon transcripts for post-transcriptional silencing and/or target transposon DNA for repressive chromatin modification and DNA methylation. These properties produce robust systems that limit the catastrophic consequences of transposon mobilization, which can result in the accumulation of deleterious mutations, changes in gene expression patterns, and conditions such as gonadal hypotrophy and sterility.
Figures


References
-
- Kazazian HH., Jr Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. - PubMed
-
- Kim JM, et al. Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 1998;8:464–478. - PubMed
-
- Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
-
- Flavell RB, et al. Genome size and the proportion of repeated nucleotide sequence DNA in plants. Biochem. Genet. 1974;12:257–269. - PubMed
-
- Kapitonov VV, Jurka J. Helitrons on a roll: eukaryotic rolling-circle transposons. Trends Genet. 2007;23:521–529. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

