Examining the link between chromosomal instability and aneuploidy in human cells
- PMID: 18283116
- PMCID: PMC2265570
- DOI: 10.1083/jcb.200712029
Examining the link between chromosomal instability and aneuploidy in human cells
Abstract
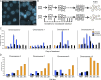
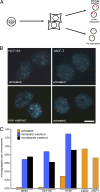
Solid tumors can be highly aneuploid and many display high rates of chromosome missegregation in a phenomenon called chromosomal instability (CIN). In principle, aneuploidy is the consequence of CIN, but the relationship between CIN and aneuploidy has not been clearly defined. In this study, we use live cell imaging and clonal cell analyses to evaluate the fidelity of chromosome segregation in chromosomally stable and unstable human cells. We show that improper microtubule-chromosome attachment (merotely) is a cause of chromosome missegregation in unstable cells and that increasing chromosome missegregation rates by elevating merotely during consecutive mitoses generates CIN in otherwise stable, near-diploid cells. However, chromosome missegregation compromises the proliferation of diploid cells, indicating that phenotypic changes that permit the propagation of nondiploid cells must combine with elevated chromosome missegregation rates to generate aneuploid cells with CIN.
Figures


Comment in
-
The wages of CIN.J Cell Biol. 2008 Feb 25;180(4):661-3. doi: 10.1083/jcb.200801030. Epub 2008 Feb 18. J Cell Biol. 2008. PMID: 18283117 Free PMC article. Review.
References
-
- Cahill, D.P., C. Lengauer, J. Yu, G.J. Riggins, J.K. Wilson, S.D. Markowitz, K.W. Kinzler, and B. Vogelstein. 1998. Mutations of mitotic checkpoint genes in human cancers. Nature. 392:300–303. - PubMed
-
- Cimini, D., and F. Degrassi. 2005. Aneuploidy: a matter of bad connections. Trends Cell Biol. 15:442–451. - PubMed
-
- Cimini, D., C. Tanzarella, and F. Degrassi. 1999. Differences in malsegregation rates obtained by scoring ana-telophases or binucleate cells. Mutagenesis. 14:563–568. - PubMed
-
- Cimini, D., L.A. Cameron, and E.D. Salmon. 2004. Anaphase spindle mechanics prevent mis-segregation of merotelically oriented chromosomes. Curr. Biol. 14:2149–2155. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

