Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development
- PMID: 18284664
- PMCID: PMC2265709
- DOI: 10.1186/1749-8104-3-5
Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development
Abstract
Background: In the mammalian brain, neural stem cells divide asymmetrically and often amplify the number of progeny they generate via symmetrically dividing intermediate progenitors. Here we investigate whether specific neural stem cell-like neuroblasts in the brain of Drosophila might also amplify neuronal proliferation by generating symmetrically dividing intermediate progenitors.
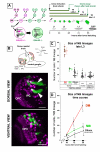
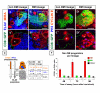
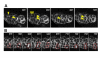
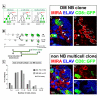
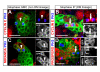
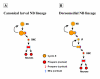
Results: Cell lineage-tracing and genetic marker analysis show that remarkably large neuroblast lineages exist in the dorsomedial larval brain of Drosophila. These lineages are generated by brain neuroblasts that divide asymmetrically to self renew but, unlike other brain neuroblasts, do not segregate the differentiating cell fate determinant Prospero to their smaller daughter cells. These daughter cells continue to express neuroblast-specific molecular markers and divide repeatedly to produce neural progeny, demonstrating that they are proliferating intermediate progenitors. The proliferative divisions of these intermediate progenitors have novel cellular and molecular features; they are morphologically symmetrical, but molecularly asymmetrical in that key differentiating cell fate determinants are segregated into only one of the two daughter cells.
Conclusion: Our findings provide cellular and molecular evidence for a new mode of neurogenesis in the larval brain of Drosophila that involves the amplification of neuroblast proliferation through intermediate progenitors. This type of neurogenesis bears remarkable similarities to neurogenesis in the mammalian brain, where neural stem cells as primary progenitors amplify the number of progeny they generate through generation of secondary progenitors. This suggests that key aspects of neural stem cell biology might be conserved in brain development of insects and mammals.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

