Amplification of multiple genomic loci from single cells isolated by laser micro-dissection of tissues
- PMID: 18284708
- PMCID: PMC2266725
- DOI: 10.1186/1472-6750-8-17
Amplification of multiple genomic loci from single cells isolated by laser micro-dissection of tissues
Abstract
Background: Whole genome amplification (WGA) and laser assisted micro-dissection represent two recently developed technologies that can greatly advance biological and medical research. WGA allows the analysis of multiple genomic loci from a single genome and has been performed on single cells from cell suspensions and from enzymatically-digested tissues. Laser micro-dissection makes it possible to isolate specific single cells from heterogeneous tissues.
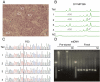
Results: Here we applied for the first time WGA on laser micro-dissected single cells from stained tissue sections, and developed a protocol for sequentially performing the two procedures. The combined procedure allows correlating the cell's genome with its natural morphology and precise anatomical position. From each cell we amplified 122 genomic and mitochondrial loci. In cells obtained from fresh tissue sections, 64.5% of alleles successfully amplified to approximately 700000 copies each, and mitochondrial DNA was amplified successfully in all cells. Multiplex PCR amplification and analysis of cells from pre-stored sections yielded significantly poorer results. Sequencing and capillary electrophoresis of WGA products allowed detection of slippage mutations in microsatellites (MS), and point mutations in P53.
Conclusion: Comprehensive genomic analysis of single cells from stained tissue sections opens new research opportunities for cell lineage and depth analyses, genome-wide mutation surveys, and other single cell assays.
Figures

References
-
- Kobashi H, Yaoi T, Oda R, Okajima S, Fujiwara H, Kubo T, Fushiki S. Lysophospholipid receptors are differentially expressed in rat terminal schwann cells, as revealed by a single cell rt-PCR and in situ hybridization. Acta Histochem Cytochem. 2006;39:55–60. doi: 10.1267/ahc.06002. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

