Human branch point consensus sequence is yUnAy
- PMID: 18285363
- PMCID: PMC2367711
- DOI: 10.1093/nar/gkn073
Human branch point consensus sequence is yUnAy
Abstract
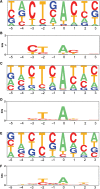
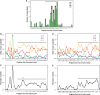
Yeast carries a strictly conserved branch point sequence (BPS) of UACUAAC, whereas the human BPS is degenerative and is less well characterized. The human consensus BPS has never been extensively explored in vitro to date. Here, we sequenced 367 clones of lariat RT-PCR products arising from 52 introns of 20 human housekeeping genes. Among the 367 clones, a misincorporated nucleotide at the branch point was observed in 181 clones, for which we can precisely pinpoint the branch point. The branch points were comprised of 92.3% A, 3.3% C, 1.7% G and 2.8% U. Our analysis revealed that the human consensus BPS is simply yUnAy, where the underlined is the branch point at position zero and the lowercase pyrimidines ('y') are not as well conserved as the uppercase U and A. We found that the branch points are located 21-34 nucleotides upstream of the 3' end of an intron in 83% clones. We also found that the polypyrimidine tract spans 4-24 nucleotides downstream of the branch point. Our analysis demonstrates that the human BPSs are more degenerative than we have expected and that the human BPSs are likely to be recognized in combination with the polypyrimidine tract and/or the other splicing cis-elements.
Figures

References
-
- Wu S, Romfo CM, Nilsen TW, Green MR. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature. 1999;402:832–835. - PubMed
-
- Zorio DA, Blumenthal T. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature. 1999;402:835–838. - PubMed
-
- Abovich N, Rosbash M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89:403–412. - PubMed
-
- Query CC, Moore MJ, Sharp PA. Branch nucleophile selection in pre-mRNA splicing: evidence for the bulged duplex model. Genes Dev. 1994;8:587–597. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

