Interferon regulatory factor 5 (IRF5) gene variants are associated with multiple sclerosis in three distinct populations
- PMID: 18285424
- PMCID: PMC2564860
- DOI: 10.1136/jmg.2007.055012
Interferon regulatory factor 5 (IRF5) gene variants are associated with multiple sclerosis in three distinct populations
Abstract
Background: IRF5 is a transcription factor involved both in the type I interferon and the toll-like receptor signalling pathways. Previously, IRF5 has been found to be associated with systemic lupus erythematosus, rheumatoid arthritis and inflammatory bowel diseases. Here we investigated whether polymorphisms in the IRF5 gene would be associated with yet another disease with features of autoimmunity, multiple sclerosis (MS).
Methods: We genotyped nine single nucleotide polymorphisms and one insertion-deletion polymorphism in the IRF5 gene in a collection of 2337 patients with MS and 2813 controls from three populations: two case-control cohorts from Spain and Sweden, and a set of MS trio families from Finland.
Results: Two single nucleotide polymorphism (SNPs) (rs4728142, rs3807306), and a 5 bp insertion-deletion polymorphism located in the promoter and first intron of the IRF5 gene, showed association signals with values of p<0.001 when the data from all cohorts were combined. The predisposing alleles were present on the same common haplotype in all populations. Using electrophoretic mobility shift assays we observed allele specific differences in protein binding for the SNP rs4728142 and the 5 bp indel, and by a proximity ligation assay we demonstrated increased binding of the transcription factor SP1 to the risk allele of the 5 bp indel.
Conclusion: These findings add IRF5 to the short list of genes shown to be associated with MS in more than one population. Our study adds to the evidence that there might be genes or pathways that are common in multiple autoimmune diseases, and that the type I interferon system is likely to be involved in the development of these diseases.
Conflict of interest statement
Figures
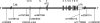
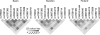
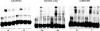

Similar articles
-
Comprehensive evaluation of the genetic variants of interferon regulatory factor 5 (IRF5) reveals a novel 5 bp length polymorphism as strong risk factor for systemic lupus erythematosus.Hum Mol Genet. 2008 Mar 15;17(6):872-81. doi: 10.1093/hmg/ddm359. Epub 2007 Dec 6. Hum Mol Genet. 2008. PMID: 18063667
-
Association of IRF5 polymorphisms with systemic lupus erythematosus in a Japanese population: support for a crucial role of intron 1 polymorphisms.Arthritis Rheum. 2008 Mar;58(3):826-34. doi: 10.1002/art.23216. Arthritis Rheum. 2008. PMID: 18311811
-
An insertion-deletion polymorphism in the interferon regulatory Factor 5 (IRF5) gene confers risk of inflammatory bowel diseases.Hum Mol Genet. 2007 Dec 15;16(24):3008-16. doi: 10.1093/hmg/ddm259. Epub 2007 Sep 19. Hum Mol Genet. 2007. PMID: 17881657
-
Association of the IRF5 rs2070197 polymorphism with systemic lupus erythematosus: a meta-analysis.Clin Rheumatol. 2015 Sep;34(9):1495-501. doi: 10.1007/s10067-015-3036-5. Epub 2015 Aug 2. Clin Rheumatol. 2015. PMID: 26233721 Review.
-
Association of IRF5, STAT4 and BLK with systemic lupus erythematosus and other rheumatic diseases.Nihon Rinsho Meneki Gakkai Kaishi. 2010;33(2):57-65. doi: 10.2177/jsci.33.57. Nihon Rinsho Meneki Gakkai Kaishi. 2010. PMID: 20453440 Review.
Cited by
-
Sample reproducibility of genetic association using different multimarker TDTs in genome-wide association studies: characterization and a new approach.PLoS One. 2012;7(2):e29613. doi: 10.1371/journal.pone.0029613. Epub 2012 Feb 17. PLoS One. 2012. PMID: 22363405 Free PMC article.
-
Spontaneous mutation of the Dock2 gene in Irf5-/- mice complicates interpretation of type I interferon production and antibody responses.Proc Natl Acad Sci U S A. 2012 Apr 10;109(15):E898-904. doi: 10.1073/pnas.1118155109. Epub 2012 Mar 19. Proc Natl Acad Sci U S A. 2012. PMID: 22431588 Free PMC article.
-
Microglia regulate myelin clearance and cholesterol metabolism after demyelination via interferon regulatory factor 5.Cell Mol Life Sci. 2025 Mar 26;82(1):131. doi: 10.1007/s00018-025-05648-2. Cell Mol Life Sci. 2025. PMID: 40137979 Free PMC article.
-
Tag-SNP analysis of the GFI1-EVI5-RPL5-FAM69 risk locus for multiple sclerosis.Eur J Hum Genet. 2010 Jul;18(7):827-31. doi: 10.1038/ejhg.2009.240. Epub 2010 Jan 20. Eur J Hum Genet. 2010. PMID: 20087403 Free PMC article.
-
Association of interferon regulatory factor 5 (IRF5) gene polymorphisms with juvenile idiopathic arthritis.Clin Rheumatol. 2018 Oct;37(10):2661-2665. doi: 10.1007/s10067-018-4010-9. Epub 2018 Feb 8. Clin Rheumatol. 2018. PMID: 29423720
References
-
- Davidson A, Diamond B. Autoimmune diseases. N Engl J Med 2001;345:340–50 - PubMed
-
- Kalman B, Lublin FD. The genetics of multiple sclerosis. A review. Biomed Pharmacother 1999;53:358–70 - PubMed
-
- Sawcer S, Ban M, Maranian M, Yeo TW, Compston A, Kirby A, Daly MJ, De Jager PL, Walsh E, Lander ES, Rioux JD, Hafler DA, Ivinson A, Rimmler J, Gregory SG, Schmidt S, Pericak-Vance MA, Akesson E, Hillert J, Datta P, Oturai A, Ryder LP, Harbo HF, Spurkland A, Myhr KM, Laaksonen M, Booth D, Heard R, Stewart G, Lincoln R, Barcellos LF, Hauser SL, Oksenberg JR, Kenealy SJ, Haines JL. A high-density screen for linkage in multiple sclerosis. Am J Hum Genet 2005;77:454–67 - PMC - PubMed
-
- Lincoln MR, Montpetit A, Cader MZ, Saarela J, Dyment DA, Tiislar M, Ferretti V, Tienari PJ, Sadovnick AD, Peltonen L, Ebers GC, Hudson TJ. A predominant role for the HLA class II region in the association of the MHC region with multiple sclerosis. Nat Genet 2005;37:1108–12 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
