Degradation of a polyadenylated rRNA maturation by-product involves one of the three RRP6-like proteins in Arabidopsis thaliana
- PMID: 18285452
- PMCID: PMC2293077
- DOI: 10.1128/MCB.02064-07
Degradation of a polyadenylated rRNA maturation by-product involves one of the three RRP6-like proteins in Arabidopsis thaliana
Abstract
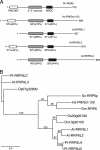
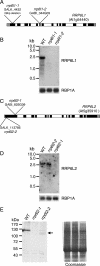
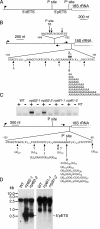
Yeast Rrp6p and its human counterpart, PM/Scl100, are exosome-associated proteins involved in the degradation of aberrant transcripts and processing of precursors to stable RNAs, such as the 5.8S rRNA, snRNAs, and snoRNAs. The activity of yeast Rrp6p is stimulated by the polyadenylation of its RNA substrates. We identified three RRP6-like proteins in Arabidopsis thaliana: AtRRP6L3 is restricted to the cytoplasm, whereas AtRRP6L1 and -2 have different intranuclear localizations. Both nuclear RRP6L proteins are functional, since AtRRP6L1 complements the temperature-sensitive phenotype of a yeast rrp6Delta strain and mutation of AtRRP6L2 leads to accumulation of an rRNA maturation by-product. This by-product corresponds to the excised 5' part of the 18S-5.8S-25S rRNA precursor and accumulates as a polyadenylated transcript, suggesting that RRP6L2 is involved in poly(A)-mediated RNA degradation in plant nuclei. Interestingly, the rRNA maturation by-product is a substrate of AtRRP6L2 but not of AtRRP6L1. This result and the distinctive subcellular distribution of AtRRP6L1 to -3 indicate a specialization of RRP6-like proteins in Arabidopsis.
Figures


References
-
- Alonso, J. M., A. N. Stepanova, T. J. Leisse, C. J. Kim, H. Chen, P. Shinn, D. K. Stevenson, J. Zimmerman, P. Barajas, R. Cheuk, C. Gadrinab, C. Heller, A. Jeske, E. Koesema, C. C. Meyers, H. Parker, L. Prednis, Y. Ansari, N. Choy, H. Deen, M. Geralt, N. Hazari, E. Hom, M. Karnes, C. Mulholland, R. Ndubaku, I. Schmidt, P. Guzman, L. Aguilar-Henonin, M. Schmid, D. Weigel, D. E. Carter, T. Marchand, E. Risseeuw, D. Brogden, A. Zeko, W. L. Crosby, C. C. Berry, and J. R. Ecker. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301653-657. - PubMed
-
- Andrulis, E. D., J. Werner, A. Nazarian, H. Erdjument-Bromage, P. Tempst, and J. T. Lis. 2002. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature 420837-841. - PubMed
-
- Bollenbach, T. J., G. Schuster, and D. B. Stern. 2004. Cooperation of endo- and exoribonucleases in chloroplast mRNA turnover. Prog. Nucleic Acid Res. Mol. Biol. 78305-337. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
