A glycine-arginine domain in control of the human MRE11 DNA repair protein
- PMID: 18285453
- PMCID: PMC2293076
- DOI: 10.1128/MCB.02025-07
A glycine-arginine domain in control of the human MRE11 DNA repair protein
Abstract
Human MRE11 is a key enzyme in DNA double-strand break repair and genome stability. Human MRE11 bears a glycine-arginine-rich (GAR) motif that is conserved among multicellular eukaryotic species. We investigated how this motif influences MRE11 function. Human MRE11 alone or a complex of MRE11, RAD50, and NBS1 (MRN) was methylated in insect cells, suggesting that this modification is conserved during evolution. We demonstrate that PRMT1 interacts with MRE11 but not with the MRN complex, suggesting that MRE11 arginine methylation occurs prior to the binding of NBS1 and RAD50. Moreover, the first six methylated arginines are essential for the regulation of MRE11 DNA binding and nuclease activity. The inhibition of arginine methylation leads to a reduction in MRE11 and RAD51 focus formation on a unique double-strand break in vivo. Furthermore, the MRE11-methylated GAR domain is sufficient for its targeting to DNA damage foci and colocalization with gamma-H2AX. These studies highlight an important role for the GAR domain in regulating MRE11 function at the biochemical and cellular levels during DNA double-strand break repair.
Figures


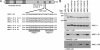
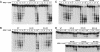




References
-
- Adams, M. M., B. Wang, Z. Xia, J. C. Morales, X. Lu, L. A. Donehower, D. A. Bochar, S. J. Elledge, and P. B. Carpenter. 2005. 53BP1 oligomerization is independent of its methylation by PRMT1. Cell Cycle 41854-1861. - PubMed
-
- Bedford, M. T., and S. Richard. 2005. Arginine methylation an emerging regulator of protein function. Mol. Cell 18263-272. - PubMed
-
- Boisvert, F. M., J. Cote, M. C. Boulanger, and S. Richard. 2003. A proteomic analysis of arginine-methylated protein complexes. Mol. Cell. Proteomics 21319-1330. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
