Ezh2 requires PHF1 to efficiently catalyze H3 lysine 27 trimethylation in vivo
- PMID: 18285464
- PMCID: PMC2293112
- DOI: 10.1128/MCB.02017-07
Ezh2 requires PHF1 to efficiently catalyze H3 lysine 27 trimethylation in vivo
Abstract
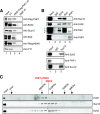
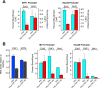
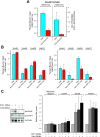
The mammalian Polycomblike protein PHF1 was previously shown to interact with the Polycomb group (PcG) protein Ezh2, a histone methyltransferase whose activity is pivotal in sustaining gene repression during development and in adulthood. As Ezh2 is active only when part of the Polycomb Repressive Complexes (PRC2-PRC4), we examined the functional role of its interaction with PHF1. Chromatin immunoprecipitation experiments revealed that PHF1 resides along with Ezh2 at Ezh2-regulated genes such as the HoxA loci and the non-Hox MYT1 and WNT1 genes. Knockdown of PHF1 or of Ezh2 led to up-regulated HoxA gene expression. Interestingly, depletion of PHF1 did correlate with reduced occupancy of Bmi-1, a PRC1 component. As expected, knockdown of Ezh2 led to reduced levels of its catalytic products H3K27me2/H3K27me3. However, reduced levels of PHF1 also led to decreased global levels of H3K27me3. Notably, the levels of H3K27me3 decreased while those of H3K27me2 increased at the up-regulated HoxA loci tested. Consistent with this, the addition of PHF1 specifically stimulated the ability of Ezh2 to catalyze H3K27me3 but not H3K27me1/H3K27me2 in vitro. We conclude that PHF1 modulates the activity of Ezh2 in favor of the repressive H3K27me3 mark. Thus, we propose that PHF1 is a determinant in PcG-mediated gene repression.
Figures




Similar articles
-
Role of hPHF1 in H3K27 methylation and Hox gene silencing.Mol Cell Biol. 2008 Mar;28(5):1862-72. doi: 10.1128/MCB.01589-07. Epub 2007 Dec 17. Mol Cell Biol. 2008. PMID: 18086877 Free PMC article.
-
Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27).Proc Natl Acad Sci U S A. 2012 Feb 21;109(8):2989-94. doi: 10.1073/pnas.1116418109. Epub 2012 Feb 8. Proc Natl Acad Sci U S A. 2012. PMID: 22323599 Free PMC article.
-
A model for transmission of the H3K27me3 epigenetic mark.Nat Cell Biol. 2008 Nov;10(11):1291-300. doi: 10.1038/ncb1787. Epub 2008 Oct 19. Nat Cell Biol. 2008. PMID: 18931660
-
EZH2 methyltransferase and H3K27 methylation in breast cancer.Int J Biol Sci. 2012;8(1):59-65. doi: 10.7150/ijbs.8.59. Epub 2011 Nov 18. Int J Biol Sci. 2012. PMID: 22211105 Free PMC article. Review.
-
Aberrations of EZH2 in cancer.Clin Cancer Res. 2011 May 1;17(9):2613-8. doi: 10.1158/1078-0432.CCR-10-2156. Epub 2011 Mar 2. Clin Cancer Res. 2011. PMID: 21367748 Review.
Cited by
-
Sound of silence: the properties and functions of repressive Lys methyltransferases.Nat Rev Mol Cell Biol. 2015 Aug;16(8):499-513. doi: 10.1038/nrm4029. Nat Rev Mol Cell Biol. 2015. PMID: 26204160 Review.
-
Histone H3.3 phosphorylation amplifies stimulation-induced transcription.Nature. 2020 Jul;583(7818):852-857. doi: 10.1038/s41586-020-2533-0. Epub 2020 Jul 22. Nature. 2020. PMID: 32699416 Free PMC article.
-
Nuclear organization and genome function.Annu Rev Cell Dev Biol. 2012;28:163-87. doi: 10.1146/annurev-cellbio-101011-155824. Epub 2012 Aug 17. Annu Rev Cell Dev Biol. 2012. PMID: 22905954 Free PMC article. Review.
-
The Bmi-1 polycomb protein antagonizes the (-)-epigallocatechin-3-gallate-dependent suppression of skin cancer cell survival.Carcinogenesis. 2010 Mar;31(3):496-503. doi: 10.1093/carcin/bgp314. Epub 2009 Dec 16. Carcinogenesis. 2010. PMID: 20015867 Free PMC article.
-
Polycomb repressor complex 2 regulates HOXA9 and HOXA10, activating ID2 in NK/T-cell lines.Mol Cancer. 2010 Jun 17;9:151. doi: 10.1186/1476-4598-9-151. Mol Cancer. 2010. PMID: 20565746 Free PMC article.
References
-
- Boyer, L. A., K. Plath, J. Zeitlinger, T. Brambrink, L. A. Medeiros, T. I. Lee, S. S. Levine, M. Wernig, A. Tajonar, M. K. Ray, G. W. Bell, A. P. Otte, M. Vidal, D. K. Gifford, R. A. Young, and R. Jaenisch. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441349-353. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
