The hepatitis C virus replicon presents a higher barrier to resistance to nucleoside analogs than to nonnucleoside polymerase or protease inhibitors
- PMID: 18285474
- PMCID: PMC2346640
- DOI: 10.1128/AAC.01317-07
The hepatitis C virus replicon presents a higher barrier to resistance to nucleoside analogs than to nonnucleoside polymerase or protease inhibitors
Abstract
Specific inhibitors of hepatitis C virus (HCV) replication that target the NS3/4A protease (e.g., VX-950) or the NS5B polymerase (e.g., R1479/R1626, PSI-6130/R7128, NM107/NM283, and HCV-796) have advanced into clinical development. Treatment of patients with VX-950 or HCV-796 rapidly selected for drug-resistant variants after a 14-day monotherapy treatment period. However, no viral resistance was identified after monotherapy with R1626 (prodrug of R1479) or NM283 (prodrug of NM107) after 14 days of monotherapy. Based upon the rapid selection of resistance to the protease and nonnucleoside inhibitors during clinical trials and the lack of selection of resistance to the nucleoside inhibitors, we used the replicon system to determine whether nucleoside inhibitors demonstrate a higher genetic barrier to resistance than protease and nonnucleoside inhibitors. Treatment of replicon cells with nucleoside inhibitors at 10 and 15 times the 50% effective concentration resulted in clearance of the replicon, while treatment with a nonnucleoside or protease inhibitor selected resistant colonies. In combination, the presence of a nucleoside inhibitor reduced the frequency of colonies resistant to the other classes of inhibitors. These results indicate that the HCV replicon presents a higher barrier to the selection of resistance to nucleoside inhibitors than to nonnucleoside or protease inhibitors. Furthermore, the combination of a nonnucleoside or protease inhibitor with a nucleoside polymerase inhibitor could have a clear clinical benefit through the delay of resistance emergence.
Figures



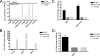

References
-
- Anonymous. 1997. Hepatitis C: global prevalence. Wkly. Epidemiol. Rec. 72:341-344. - PubMed
-
- Armstrong, G. L., A. Wasley, E. P. Simard, G. M. McQuillan, W. L. Kuhnert, and M. J. Alter. 2006. The prevalence of hepatitis C virus infection in the United States, 1999 through 2002. Ann. Intern. Med. 144:705-714. - PubMed
-
- Chandra, P., D. Raible, D. Harper, J. Speth, S. Villano, and G. Bichier. 2006. Antiviral activity of the non nucleoside polymerase inhibitor, HCV-796, in patients with chronic hepatitis C virus: preliminary results from a randomized, double-blind, placebo-controlled, ascending multiple dose study. Gastroenterology 130:A-748.
-
- De Francesco, R., and G. Migliaccio. 2005. Challenges and successes in developing new therapies for hepatitis C. Nature 436:953-960. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

