Quality control of DNA break metabolism: in the 'end', it's a good thing
- PMID: 18285819
- PMCID: PMC2262039
- DOI: 10.1038/emboj.2008.11
Quality control of DNA break metabolism: in the 'end', it's a good thing
Abstract
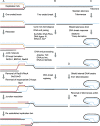
DNA ends pose specific problems in the control of genetic information quality. Ends of broken DNA need to be rejoined to avoid genome rearrangements, whereas natural DNA ends of linear chromosomes, telomeres, need to be stable and hidden from the DNA damage response. Efficient DNA end metabolism, either at induced DNA breaks or telomeres, does not result from the machine-like precision of molecular reactions, but rather from messier, more stochastic processes. The necessary molecular interactions are dynamically unstable, with constructive and destructive processes occurring in competition. In the end, quality control comes from the constant building up and tearing down of inappropriate, but also appropriate reaction steps in combination with factors that only slightly shift the equilibrium to eventually favour appropriate events. Thus, paradoxically, enzymes antagonizing DNA end metabolism help to ensure that genome maintenance becomes a robust process.
Figures
References
-
- Agarwal S, Tafel AA, Kanaar R (2006) DNA double-strand break repair and chromosome translocations. DNA Repair (Amst) 5: 1075–1081 - PubMed
-
- Amitani I, Baskin RJ, Kowalczykowski SC (2006) Visualization of Rad54, a chromatin remodeling protein, translocating on single DNA molecules. Mol Cell 23: 143–148 - PubMed
-
- Boule JB, Vega LR, Zakian VA (2005) The yeast Pif1p helicase removes telomerase from telomeric DNA. Nature 438: 57–61 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

