Molecular pathways involved in loss of kidney graft function with tubular atrophy and interstitial fibrosis
- PMID: 18286166
- PMCID: PMC2242778
- DOI: 10.2119/2007-00111.Maluf
Molecular pathways involved in loss of kidney graft function with tubular atrophy and interstitial fibrosis
Abstract
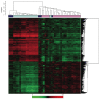
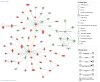
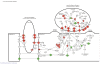
Loss of kidney graft function with tubular atrophy (TA) and interstitial fibrosis (IF) causes most kidney allograft losses. We aimed to identify the molecular pathways involved in IF/TA progression. Kidney biopsies from normal kidneys (n = 24), normal allografts (n = 6), and allografts with IF/TA (n = 17) were analyzed using high-density oligonucleotide microarray. Probe set level tests of hypotheses tests were conducted to identify genes with a significant trend in gene expression across the three groups using Jonckheere-Terpstra test for trend. Interaction networks and functional analysis were used. An unsupervised hierarchical clustering analysis showed that all the IF/TA samples were associated with high correlation. Gene ontology classified the differentially expressed genes as related to immune response, inflammation, and matrix deposition. Chemokines (CX), CX receptor (for example, CCL5 and CXCR4), interleukin, and interleukin receptor (for example, IL-8 and IL10RA) genes were overexpressed in IF/TA samples compared with normal allografts and normal kidneys. Genes involved in apoptosis (for example, CASP4 and CASP5) were importantly overexpressed in IF/TA. Genes related to angiogenesis (for example, ANGPTL3, ANGPT2, and VEGF) were downregulated in IF/TA. Genes related to matrix production-deposition were upregulated in IF/TA. A distinctive gene expression pattern was observed in IF/TA samples compared with normal allografts and normal kidneys. We were able to establish a trend in gene expression for genes involved in different pathways among the studied groups. The top-scored networks were related to immune response, inflammation, and cell-to-cell interaction, showing the importance of chronic inflammation in progressive graft deterioration.
Figures


References
-
- Hariharan S, et al. Improved graft survival after renal transplantation in the United States, 1988 to 1996. N Engl J Med. 2000;342:605–12. - PubMed
-
- Meier-Kriesche HU, et al. Lack of improvement in renal allograft survival despite a marked decrease in acute rejection rates over the most recent era. Am J Transplant. 2004;4:378–83. - PubMed
-
- Solez K, et al. Banff ‘05 Meeting Report: differential diagnosis of chronic allograft injury and elimination of chronic allograft nephropathy (‘CAN’) Am J Transplant. 2007;7:518–26. - PubMed
-
- Halloran PF, et al. Assessing long-term nephron loss: is it time to kick the CAN grading system? Am J Transplant. 2004;11:1729–30. - PubMed
-
- Nankivell BJ, Chapman JR. Chronic allograft nephropathy: current concepts and future directions. Transplantation. 2006;81:643–54. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

