Ybp2 associates with the central kinetochore of Saccharomyces cerevisiae and mediates proper mitotic progression
- PMID: 18286174
- PMCID: PMC2238814
- DOI: 10.1371/journal.pone.0001617
Ybp2 associates with the central kinetochore of Saccharomyces cerevisiae and mediates proper mitotic progression
Abstract
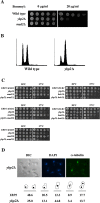
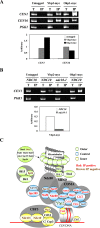
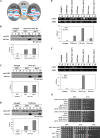
The spindle checkpoint ensures the accurate segregation of chromosomes by monitoring the status of kinetochore attachment to microtubules. Simultaneous mutations in one of several kinetochore and cohesion genes and a spindle checkpoint gene cause a synthetic-lethal or synthetic-sick phenotype. A synthetic genetic array (SGA) analysis using a mad2Delta query mutant strain of yeast identified YBP2, a gene whose product shares sequence similarity with the product of YBP1, which is required for H(2)O(2)-induced oxidation of the transcription factor Yap1. ybp2Delta was sensitive to benomyl and accumulated at the mitotic stage of the cell cycle. Ybp2 physically associates with proteins of the COMA complex (Ctf19, Okp1, Mcm21, and Ame1) and 3 components of the Ndc80 complex (Ndc80, Nuf2, and Spc25 but not Spc24) in the central kinetochore and with Cse4 (the centromeric histone and CENP-A homolog). Chromatin-immunoprecipitation analyses revealed that Ybp2 associates specifically with CEN DNA. Furthermore, ybp2Delta showed synthetic-sick interactions with mutants of the genes that encode the COMA complex components. Ybp2 seems to be part of a macromolecular kinetochore complex and appears to contribute to the proper associations among the central kinetochore subcomplexes and the kinetochore-specific nucleosome.
Conflict of interest statement
Figures


References
-
- Biggins S, Walczak CE. Captivating capture: how microtubules attach to kinetochores. Curr Biol. 2003;13:R449–460. - PubMed
-
- Kitagawa K, Hieter P. Evolutionary conservation between budding yeast and human kinetochores. Nat Rev Mol Cell Biol. 2001;2:678–687. - PubMed
-
- Goshima G, Yanagida M. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell. 2000;100:619–633. - PubMed
-
- He X, Asthana S, Sorger PK. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell. 2000;101:763–775. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

