Sequence evidence in the archaeal genomes that tRNAs emerged through the combination of ancestral genes as 5' and 3' tRNA halves
- PMID: 18286179
- PMCID: PMC2237900
- DOI: 10.1371/journal.pone.0001622
Sequence evidence in the archaeal genomes that tRNAs emerged through the combination of ancestral genes as 5' and 3' tRNA halves
Abstract
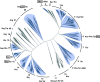
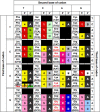
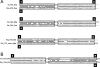
The discovery of separate 5' and 3' halves of transfer RNA (tRNA) molecules-so-called split tRNA-in the archaeal parasite Nanoarchaeum equitans made us wonder whether ancestral tRNA was encoded on 1 or 2 genes. We performed a comprehensive phylogenetic analysis of tRNAs in 45 archaeal species to explore the relationship between the three types of tRNAs (nonintronic, intronic and split). We classified 1953 mature tRNA sequences into 22 clusters. All split tRNAs have shown phylogenetic relationships with other tRNAs possessing the same anticodon. We also mimicked split tRNA by artificially separating the tRNA sequences of 7 primitive archaeal species at the anticodon and analyzed the sequence similarity and diversity of the 5' and 3' tRNA halves. Network analysis revealed specific characteristics of and topological differences between the 5' and 3' tRNA halves: the 5' half sequences were categorized into 6 distinct groups with a sequence similarity of >80%, while the 3' half sequences were categorized into 9 groups with a higher sequence similarity of >88%, suggesting different evolutionary backgrounds of the 2 halves. Furthermore, the combinations of 5' and 3' halves corresponded with the variation of amino acids in the codon table. We found not only universally conserved combinations of 5'-3' tRNA halves in tRNA(iMet), tRNA(Thr), tRNA(Ile), tRNA(Gly), tRNA(Gln), tRNA(Glu), tRNA(Asp), tRNA(Lys), tRNA(Arg) and tRNA(Leu) but also phylum-specific combinations in tRNA(Pro), tRNA(Ala), and tRNA(Trp). Our results support the idea that tRNA emerged through the combination of separate genes and explain the sequence diversity that arose during archaeal tRNA evolution.
Conflict of interest statement
Figures



References
-
- Weiner AM, Maizels N. The genomic tag hypothesis: modern viruses as molecular fossils of ancient strategies for genomic replication, and clues regarding the origin of protein synthesis. Biol Bull. 1999;196:327–328. discussion 329-30. - PubMed
-
- Di Giulio M. The non-monophyletic origin of the tRNA molecule and the origin of genes only after the evolutionary stage of the last universal common ancestor (LUCA). J Theor Biol. 2006;240:343–352. - PubMed
-
- Nagaswamy U, Fox GE. RNA ligation and the origin of tRNA. Orig Life Evol Biosph. 2003;33:199–209. - PubMed
-
- Di Giulio M. The non-monophyletic origin of the tRNA molecule. J Theor Biol. 1999;197:403–414. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

