MicroRNAs and the advent of vertebrate morphological complexity
- PMID: 18287013
- PMCID: PMC2268565
- DOI: 10.1073/pnas.0712259105
MicroRNAs and the advent of vertebrate morphological complexity
Abstract
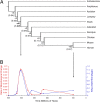
The causal basis of vertebrate complexity has been sought in genome duplication events (GDEs) that occurred during the emergence of vertebrates, but evidence beyond coincidence is wanting. MicroRNAs (miRNAs) have recently been identified as a viable causal factor in increasing organismal complexity through the action of these approximately 22-nt noncoding RNAs in regulating gene expression. Because miRNAs are continuously being added to animalian genomes, and, once integrated into a gene regulatory network, are strongly conserved in primary sequence and rarely secondarily lost, their evolutionary history can be accurately reconstructed. Here, using a combination of Northern analyses and genomic searches, we show that 41 miRNA families evolved at the base of Vertebrata, as they are found and/or detected in lamprey, but not in either ascidians or amphioxus (or any other nonchordate taxon). When placed into temporal context, the rate of miRNA acquisition and the extent of phenotypic evolution are anomalously high early in vertebrate history, far outstripping any other episode in chordate evolution. The genomic position of miRNA paralogues in humans, together with gene trees incorporating lamprey orthologues, indicates that although GDEs can account for an increase in the diversity of miRNA family members, which occurred before the last common ancestor of all living vertebrates, GDEs cannot account for the origin of these novel families themselves. We hypothesize that lying behind the origin of vertebrate complexity is the dramatic expansion of the noncoding RNA inventory including miRNAs, rather than an increase in the protein-encoding inventory caused by GDEs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

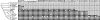

References
-
- Valentine JW, Collins AG, Meyer CP. Morphological complexity increase in metazoans. Paleobiology. 1994;20:131–142.
-
- Aburomia R, Khaner O, Sidow A. Functional evolution in the ancestral lineage of vertebrates or when genomic complexity was wagging its morphological tail. J Struct Funct Genomics. 2003;3:45–52. - PubMed
-
- Ohno S. Evolution by Gene Duplication. Heidelberg: Springer; 1970.
-
- Holland PWH, Garcia-Fernàndez J, Williams, Nic A, Sidow A. In: The Evolution of Developmental Regulatory Mechanisms. Akam M, Holland P, Ingham P, Wray G, editors. Cambridge, UK: The Company of Biologists Limited; 1994. pp. 125–133.
-
- Sidow A. Genome duplications in the evolution of early vertebrates. Curr Opin Genet Dev. 1996;6:715–722. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

