Collagen fibril architecture, domain organization, and triple-helical conformation govern its proteolysis
- PMID: 18287018
- PMCID: PMC2268544
- DOI: 10.1073/pnas.0710588105
Collagen fibril architecture, domain organization, and triple-helical conformation govern its proteolysis
Abstract
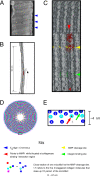
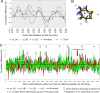
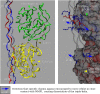
We describe the molecular structure of the collagen fibril and how it affects collagen proteolysis or "collagenolysis." The fibril-forming collagens are major components of all mammalian connective tissues, providing the structural and organizational framework for skin, blood vessels, bone, tendon, and other tissues. The triple helix of the collagen molecule is resistant to most proteinases, and the matrix metalloproteinases that do proteolyze collagen are affected by the architecture of collagen fibrils, which are notably more resistant to collagenolysis than lone collagen monomers. Until now, there has been no molecular explanation for this. Full or limited proteolysis of the collagen fibril is known to be a key process in normal growth, development, repair, and cell differentiation, and in cancerous tumor progression and heart disease. Peptide fragments generated by collagenolysis, and the conformation of exposed sites on the fibril as a result of limited proteolysis, regulate these processes and that of cellular attachment, but it is not known how or why. Using computational and molecular visualization methods, we found that the arrangement of collagen monomers in the fibril (its architecture) protects areas vulnerable to collagenolysis and strictly governs the process. This in turn affects the accessibility of a cell interaction site located near the cleavage region. Our observations suggest that the C-terminal telopeptide must be proteolyzed before collagenase can gain access to the cleavage site. Collagenase then binds to the substrate's "interaction domain," which facilitates the triple-helix unwinding/dissociation function of the enzyme before collagenolysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Tam EM, Wu YI, Butler GS, Stack MS, Overall CM. Collagen binding properties of the membrane type 1 matrix metalloproteinase (MT1-MMP) hemopexin C domain. The ectodomain of the 44-kDa autocatalytic product of MT1-MMP inhibits cell invasion by disrupting native type I collagen cleavage. J Biol Chem. 2002;277:39005–39014. - PubMed
-
- Rich A, Crick FH. The molecular structure of collagen. J Mol Biol. 1961;3:483–506. - PubMed
-
- Okuyama K, Takayanagi M, Ashida T, Kakudo M. A new structural model for collagen. Polymer J. 1971;9:341–343.
-
- Kramer RZ, Bella J, Brodsky B, Berman HM. The crystal and molecular structure of a collagen-like peptide with a biologically relevant sequence. J Mol Biol. 2001;311:131–147. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

