Dynamics of Pseudomonas aeruginosa genome evolution
- PMID: 18287045
- PMCID: PMC2268591
- DOI: 10.1073/pnas.0711982105
Dynamics of Pseudomonas aeruginosa genome evolution
Abstract
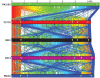
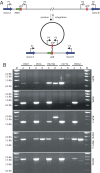
One of the hallmarks of the Gram-negative bacterium Pseudomonas aeruginosa is its ability to thrive in diverse environments that includes humans with a variety of debilitating diseases or immune deficiencies. Here we report the complete sequence and comparative analysis of the genomes of two representative P. aeruginosa strains isolated from cystic fibrosis (CF) patients whose genetic disorder predisposes them to infections by this pathogen. The comparison of the genomes of the two CF strains with those of other P. aeruginosa presents a picture of a mosaic genome, consisting of a conserved core component, interrupted in each strain by combinations of specific blocks of genes. These strain-specific segments of the genome are found in limited chromosomal locations, referred to as regions of genomic plasticity. The ability of P. aeruginosa to shape its genomic composition to favor survival in the widest range of environmental reservoirs, with corresponding enhancement of its metabolic capacity is supported by the identification of a genomic island in one of the sequenced CF isolates, encoding enzymes capable of degrading terpenoids produced by trees. This work suggests that niche adaptation is a major evolutionary force influencing the composition of bacterial genomes. Unlike genome reduction seen in host-adapted bacterial pathogens, the genetic capacity of P. aeruginosa is determined by the ability of individual strains to acquire or discard genomic segments, giving rise to strains with customized genomic repertoires. Consequently, this organism can survive in a wide range of environmental reservoirs that can serve as sources of the infecting organisms.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Tan MW. Cross-species infections and their analysis. Annu Rev Microbiol. 2002;56:539–565. - PubMed
-
- Moran NA. Microbial minimalism: Genome reduction in bacterial pathogens. Cell. 2002;108:583–586. - PubMed
-
- Elting LS, Rubenstein EB, Rolston KV, Bodey GP. Outcomes of bacteremia in patients with cancer and neutropenia: Observations from two decades of epidemiological and clinical trials. Clin Infect Dis. 1997;25:247–259. - PubMed
-
- Diaz E, Munoz E, Agbaht K, Rello J. Management of ventilator-associated pneumonia caused by multiresistant bacteria. Curr Opin Crit Care. 2007;13:45–50. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

