DNA-binding study identifies C-box and hybrid C/G-box or C/A-box motifs as high-affinity binding sites for STF1 and LONG HYPOCOTYL5 proteins
- PMID: 18287490
- PMCID: PMC2287355
- DOI: 10.1104/pp.107.113217
DNA-binding study identifies C-box and hybrid C/G-box or C/A-box motifs as high-affinity binding sites for STF1 and LONG HYPOCOTYL5 proteins
Abstract
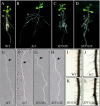
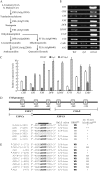
LONG HYPOCOTYL5 (HY5) is a bZIP (basic leucine zipper) transcription factor that activates photomorphogenesis and root development in Arabidopsis (Arabidopsis thaliana). Previously, STF1 (soybean [Glycine max] TGACG-motif binding factor 1), a homologous legume protein with a RING-finger motif and a bZIP domain, was reported in soybean. To investigate the role of STF1, the phenotypes of transgenic Arabidopsis plants overexpressing STF1 and HY5 were compared. In addition, the DNA-binding properties of STF1 and HY5 were extensively studied using random binding site selection and electrophoretic mobility shift assay. Overexpression of STF1 in the hy5 mutant of Arabidopsis restored wild-type photomorphogenic and root development phenotypes of short hypocotyl, accumulation of chlorophyll, and root gravitropism with partial restoration of anthocyanin accumulation. This supports that STF1 is a homolog of HY5 with a role in light and hormone signaling. The DNA-binding properties of STF1 and HY5 are shown to be similar to each other in recognizing many ACGT-containing elements with a consensus sequence motif of 5'-(G/A)(G/A) TGACGT(C/G/A)(A/T/G)-3'. The motif represents a characteristically strong preference for flanking sequence to TGACGT and a larger sequence than the sequences recognized by the G-box binding factor and TGA protein families. The finding of C-box, hybrid C/G-, and C/A-boxes as high-affinity binding sites over the G-box and parameters associated with HY5 recognition define the criteria of HY5/STF1 protein-DNA interaction in the promoter regions. This study helps to predict the precise in vivo binding sites of the HY5 protein from the vast number of putative HY5 genomic binding sites analyzed by chromatin immunoprecipitation on chip.
Figures






References
-
- Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng XW (1998) Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell 1 213–222 - PubMed
-
- Cheong YH, Park JM, Yoo CM, Bahk JD, Cho MJ, Hong JC (1994) Isolation and characterization of STGA1, a member of the TGA1 family of bZIP transcription factors from soybean. Mol Cells 4 405–412
-
- Cheong YH, Yoo CM, Park JM, Ryu GR, Goekjian VH, Nagao RT, Key JL, Cho MJ, Hong JC (1998) STF1 is a novel TGACG-binding factor with a zinc-finger motif and a bZIP domain which heterodimerizes with GBF proteins. Plant J 15 199–209 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

