Protein networks supporting AP-3 function in targeting lysosomal membrane proteins
- PMID: 18287518
- PMCID: PMC2366865
- DOI: 10.1091/mbc.e08-02-0110
Protein networks supporting AP-3 function in targeting lysosomal membrane proteins
Abstract
The AP-3 adaptor complex targets selected transmembrane proteins to lysosomes and lysosome-related organelles. We reconstituted its preferred interaction with liposomes containing the ADP ribosylation factor (ARF)-1 guanosine triphosphatase (GTPase), specific cargo tails, and phosphatidylinositol-3 phosphate, and then we performed a proteomic screen to identify new proteins supporting its sorting function. We identified approximately 30 proteins belonging to three networks regulating either AP-3 coat assembly or septin polymerization or Rab7-dependent lysosomal transport. RNA interference shows that, among these proteins, the ARF-1 exchange factor brefeldin A-inhibited exchange factor 1, the ARF-1 GTPase-activating protein 1, the Cdc42-interacting Cdc42 effector protein 4, an effector of septin-polymerizing GTPases, and the phosphatidylinositol-3 kinase IIIC3 are key components regulating the targeting of lysosomal membrane proteins to lysosomes in vivo. This analysis reveals that these proteins, together with AP-3, play an essential role in protein sorting at early endosomes, thereby regulating the integrity of these organelles.
Figures
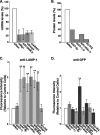
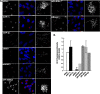
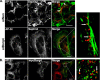
References
-
- Barral Y., Mermall V., Mooseker M. S., Snyder M. Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol. Cell. 2000;5:841–851. - PubMed
-
- Bonifacino J. S., Glick B. S. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. - PubMed
-
- Bonifacino J. S., Traub L. M. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 2003;72:395–447. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

